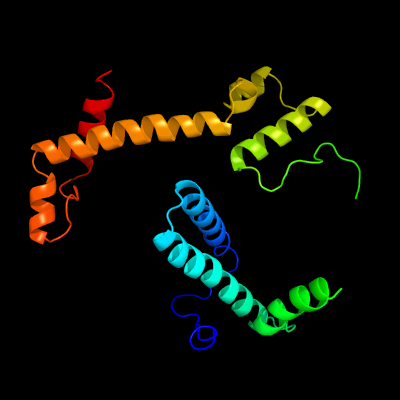
| 1 | c3crcB_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein mazg;
PDBTitle: crystal structure of escherichia coli mazg, the regulator2 of nutritional stress response
|
|
|
|
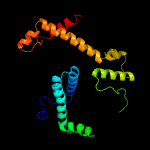
| 2 | c2yxhB_
|
|
 |
100.0 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:mazg-related protein;
PDBTitle: crystal structure of mazg-related protein from thermotoga maritima
|
|
|
|
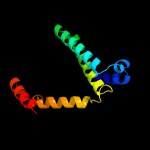
| 3 | c5n0sA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
|
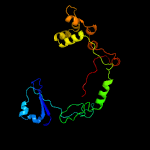
| 4 | d2a3qa1
|
|
 |
99.6 |
20 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
|
|
|
| 5 | c2q4pA_
|
|
 |
99.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein rs21-c6;
PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898
|
|
|
|
| 6 | c3obcB_
|
|
 |
99.6 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:pyrophosphatase;
PDBTitle: crystal structure of a pyrophosphatase (af1178) from archaeoglobus2 fulgidus at 1.80 a resolution
|
|
|
|
| 7 | d1vmga_
|
|
 |
98.9 |
21 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
|
|
|
| 8 | d2oiea1
|
|
 |
98.3 |
20 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
|
|
|
| 9 | d2gtad1
|
|
 |
98.2 |
22 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
|
|
|
| 10 | d2gtaa1
|
|
 |
97.7 |
22 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
|
|
|
| 11 | d1yxba1
|
|
 |
97.4 |
29 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
|
|
|
| 12 | d1y6xa1
|
|
 |
97.3 |
35 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
|
|
|
| 13 | c1yvwD_
|
|
 |
97.1 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp2 pyrophosphohydrolase from bacillus cereus. nesgc target3 bcr13.
|
|
|
|
| 14 | d1yvwa1
|
|
 |
97.1 |
17 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
|
|
|
| 15 | d2a7wa1
|
|
 |
96.9 |
24 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
|
|
|
| 16 | c2a7wF_
|
|
 |
96.9 |
24 |
PDB header:hydrolase
Chain: F: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
|
|
|
|
| 17 | c2q9lA_
|
|
 |
96.7 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of imazg from vibrio dat 722: ctag-imazg (p43212)
|
|
|
|
| 18 | c2p06A_
|
|
 |
94.5 |
37 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af_0060;
PDBTitle: crystal structure of a predicted coding region af_0060 from2 archaeoglobus fulgidus dsm 4304
|
|
|
|
| 19 | d2p06a1
|
|
 |
94.5 |
37 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:AF0060-like |
|
|
|
| 20 | c2yf3F_
|
|
 |
92.8 |
33 |
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from deinococcus2 radiodurans, complex with manganese
|
|
|
|
| 21 | c3nqwB_ |
|
not modelled |
60.5 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:cg11900;
PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
|
|
|
| 22 | c3qk9B_ |
|
not modelled |
33.6 |
23 |
PDB header:protein transport
Chain: B: PDB Molecule:mitochondrial import inner membrane translocase subunit
PDBTitle: yeast tim44 c-terminal domain complexed with cymal-3
|
|
|
| 23 | d2fxta1 |
|
not modelled |
33.4 |
20 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:TIM44-like |
|
|
| 24 | d2cw9a1 |
|
not modelled |
33.4 |
14 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:TIM44-like |
|
|
| 25 | c5h6sB_ |
|
not modelled |
33.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidase;
PDBTitle: crystal structure of hydrazidase s179a mutant complexed with a2 substrate
|
|
|
| 26 | c5jffD_ |
|
not modelled |
29.8 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:uncharacterized protein yhfg;
PDBTitle: e. coli ecfict mutant g55r in complex with ecfica
|
|
|
| 27 | c4uqfB_ |
|
not modelled |
29.4 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:gtp cyclohydrolase 1;
PDBTitle: crystal structure of listeria monocytogenes gtp cyclohydrolase i
|
|
|
| 28 | c3ah5E_ |
|
not modelled |
25.0 |
18 |
PDB header:transferase
Chain: E: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
|
|
|
| 29 | c5b3iB_ |
|
not modelled |
22.0 |
13 |
PDB header:electron transport
Chain: B: PDB Molecule:cytochrome c prime;
PDBTitle: homo-dimeric structure of cytochrome c' from thermophilic2 hydrogenophilus thermoluteolus
|
|
|
| 30 | d2j4ba1 |
|
not modelled |
21.9 |
25 |
Fold:Taf5 N-terminal domain-like
Superfamily:Taf5 N-terminal domain-like
Family:Taf5 N-terminal domain-like |
|
|
| 31 | d1e85a_ |
|
not modelled |
21.3 |
23 |
Fold:Four-helical up-and-down bundle
Superfamily:Cytochromes
Family:Cytochrome c'-like |
|
|
| 32 | c5lf9A_ |
|
not modelled |
21.0 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:nucleoside diphosphate-linked moiety x motif 22;
PDBTitle: crystal structure of human nudt22
|
|
|
| 33 | c2rfpA_ |
|
not modelled |
20.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative ntp pyrophosphohydrolase;
PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
|
|
|
| 34 | c6hn9A_ |
|
not modelled |
20.4 |
30 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:nicomicin-1;
PDBTitle: nicomicin-1 -- novel antimicrobial peptides from the arctic polychaeta2 nicomache minor provide new molecular insight into biological role of3 the brichos domain
|
|
|
| 35 | d1bpoa1 |
|
not modelled |
19.7 |
70 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Clathrin heavy-chain linker domain |
|
|
| 36 | d1cpqa_ |
|
not modelled |
19.2 |
15 |
Fold:Four-helical up-and-down bundle
Superfamily:Cytochromes
Family:Cytochrome c'-like |
|
|
| 37 | c5h6tB_ |
|
not modelled |
19.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidase;
PDBTitle: crystal structure of hydrazidase from microbacterium sp. strain hm58-2
|
|
|
| 38 | c2jz8A_ |
|
not modelled |
17.5 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
|
|
|
| 39 | c6feyF_ |
|
not modelled |
17.5 |
64 |
PDB header:peptide binding protein
Chain: F: PDB Molecule:probable insulin-like peptide 5;
PDBTitle: crystal structure of drosophila neural ectodermal development factor2 imp-l2 with drosophila dilp5 insulin
|
|
|
| 40 | c6feyH_ |
|
not modelled |
17.2 |
64 |
PDB header:peptide binding protein
Chain: H: PDB Molecule:probable insulin-like peptide 5;
PDBTitle: crystal structure of drosophila neural ectodermal development factor2 imp-l2 with drosophila dilp5 insulin
|
|
|
| 41 | c3efdK_ |
|
not modelled |
17.2 |
22 |
PDB header:immune system
Chain: K: PDB Molecule:kcsa;
PDBTitle: the crystal structure of the cytoplasmic domain of kcsa
|
|
|
| 42 | c2wfuB_ |
|
not modelled |
16.8 |
64 |
PDB header:signaling protein
Chain: B: PDB Molecule:probable insulin-like peptide 5 b chain;
PDBTitle: crystal structure of dilp5 variant db
|
|
|
| 43 | c6hqaB_ |
|
not modelled |
16.6 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:subunit (90 kda) of tfiid and saga complexes;
PDBTitle: molecular structure of promoter-bound yeast tfiid
|
|
|
| 44 | c2wfvB_ |
|
not modelled |
16.6 |
64 |
PDB header:signaling protein
Chain: B: PDB Molecule:probable insulin-like peptide 5 b chain;
PDBTitle: crystal structure of dilp5 variant c4
|
|
|
| 45 | c1wm9D_ |
|
not modelled |
16.5 |
5 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: structure of gtp cyclohydrolase i from thermus thermophilus hb8
|
|
|
| 46 | d1wura1 |
|
not modelled |
16.5 |
5 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
|
|
| 47 | c6hqaC_ |
|
not modelled |
16.3 |
26 |
PDB header:transcription
Chain: C: PDB Molecule:subunit (90 kda) of tfiid and saga complexes;
PDBTitle: molecular structure of promoter-bound yeast tfiid
|
|
|
| 48 | c4cimP_ |
|
not modelled |
15.8 |
31 |
PDB header:apoptosis
Chain: P: PDB Molecule:bcl-2-like protein 2;
PDBTitle: complex of a bcl-w bh3 mutant with a bh3 domain
|
|
|
| 49 | c4cimQ_ |
|
not modelled |
15.8 |
31 |
PDB header:apoptosis
Chain: Q: PDB Molecule:bcl-2-like protein 2;
PDBTitle: complex of a bcl-w bh3 mutant with a bh3 domain
|
|
|
| 50 | c5oezA_ |
|
not modelled |
15.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:fbp protein;
PDBTitle: crystal structure of leishmania major fructose-1,6-bisphosphatase in2 apo form.
|
|
|
| 51 | c6mzcG_ |
|
not modelled |
15.7 |
28 |
PDB header:transcription
Chain: G: PDB Molecule:transcription initiation factor tfiid subunit 5;
PDBTitle: human tfiid bc core
|
|
|
| 52 | c4b6xB_ |
|
not modelled |
15.1 |
29 |
PDB header:toxin
Chain: B: PDB Molecule:avirulence protein;
PDBTitle: crystal structure of in planta processed avrrps4
|
|
|
| 53 | d1ffgb_ |
|
not modelled |
15.0 |
56 |
Fold:Ferredoxin-like
Superfamily:CheY-binding domain of CheA
Family:CheY-binding domain of CheA |
|
|
| 54 | c1a0oH_ |
|
not modelled |
14.6 |
56 |
PDB header:chemotaxis
Chain: H: PDB Molecule:chea;
PDBTitle: chey-binding domain of chea in complex with chey
|
|
|
| 55 | c5olaD_ |
|
not modelled |
14.1 |
58 |
PDB header:transcription
Chain: D: PDB Molecule:transcription elongation factor, mitochondrial;
PDBTitle: structure of mitochondrial transcription elongation complex in complex2 with elongation factor tefm
|
|
|
| 56 | c4yj6A_ |
|
not modelled |
13.6 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:aryl acylamidase;
PDBTitle: the crystal structure of a bacterial aryl acylamidase belonging to the2 amidase signature (as) enzymes family
|
|
|
| 57 | c5k4bA_ |
|
not modelled |
13.3 |
22 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 3 subunit d;
PDBTitle: structure of eukaryotic translation initiation factor 3 subunit d2 (eif3d) cap binding domain from nasonia vitripennis, crystal form 1
|
|
|
| 58 | c2la2A_ |
|
not modelled |
13.2 |
35 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:cecropin;
PDBTitle: solution structure of papiliocin isolated from the swallowtail2 butterfly, papilio xuthus
|
|
|
| 59 | c2ctoA_ |
|
not modelled |
12.7 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:novel protein;
PDBTitle: solution structure of the hmg box like domain from human2 hypothetical protein flj14904
|
|
|
| 60 | c6a3kA_ |
|
not modelled |
12.4 |
20 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome c;
PDBTitle: crystal structure of cytochrome c' from shewanella benthica db6705
|
|
|
| 61 | c4b6xA_ |
|
not modelled |
12.3 |
29 |
PDB header:toxin
Chain: A: PDB Molecule:avirulence protein;
PDBTitle: crystal structure of in planta processed avrrps4
|
|
|
| 62 | d1mt5a_ |
|
not modelled |
11.7 |
14 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
| 63 | c1bpoA_ |
|
not modelled |
11.7 |
70 |
PDB header:membrane protein
Chain: A: PDB Molecule:protein (clathrin);
PDBTitle: clathrin heavy-chain terminal domain and linker
|
|
|
| 64 | d3d1ma1 |
|
not modelled |
11.6 |
21 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
|
|
| 65 | d2ibge1 |
|
not modelled |
11.5 |
21 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
|
|
| 66 | d1oiza1 |
|
not modelled |
11.5 |
27 |
Fold:RuvA C-terminal domain-like
Superfamily:CRAL/TRIO N-terminal domain
Family:CRAL/TRIO N-terminal domain |
|
|
| 67 | c2d1lA_ |
|
not modelled |
11.3 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:metastasis suppressor protein 1;
PDBTitle: structure of f-actin binding domain imd of mim (missing in metastasis)
|
|
|
| 68 | c5lrvA_ |
|
not modelled |
11.1 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:otu domain-containing protein 7b;
PDBTitle: structure of cezanne/otud7b otu domain bound to lys11-linked2 diubiquitin
|
|
|
| 69 | c3m1nB_ |
|
not modelled |
11.0 |
21 |
PDB header:signaling protein
Chain: B: PDB Molecule:sonic hedgehog protein;
PDBTitle: crystal structure of human sonic hedgehog n-terminal domain
|
|
|
| 70 | c4ulvB_ |
|
not modelled |
10.9 |
15 |
PDB header:electron transport
Chain: B: PDB Molecule:cytochrome c, class ii;
PDBTitle: cytochrome c prime from shewanella frigidimarina
|
|
|
| 71 | c3e4xB_ |
|
not modelled |
10.8 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:apc36150;
PDBTitle: crystal structure of putative metal-dependent hydrolases2 apc36150
|
|
|
| 72 | c3muxB_ |
|
not modelled |
10.6 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
|
|
| 73 | c5flmK_ |
|
not modelled |
10.6 |
28 |
PDB header:transcription
Chain: K: PDB Molecule:dna-directed rna polymerase ii subunit rpb11;
PDBTitle: structure of transcribing mammalian rna polymerase ii
|
|
|
| 74 | c6c6gA_ |
|
not modelled |
10.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:biuret hydrolase;
PDBTitle: an unexpected vestigial protein complex reveals the evolutionary2 origins of an s-triazine catabolic enzyme. inhibitor bound complex.
|
|
|
| 75 | c2lfcA_ |
|
not modelled |
10.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavoprotein subunit;
PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
|
|
|
| 76 | d1u5pa2 |
|
not modelled |
10.0 |
15 |
Fold:Spectrin repeat-like
Superfamily:Spectrin repeat
Family:Spectrin repeat |
|
|
| 77 | d1ej5a_ |
|
not modelled |
10.0 |
7 |
Fold:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Superfamily:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Family:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain |
|
|
| 78 | c3a2qA_ |
|
not modelled |
9.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-aminohexanoate-cyclic-dimer hydrolase;
PDBTitle: structure of 6-aminohexanoate cyclic dimer hydrolase complexed with2 substrate
|
|
|
| 79 | c3m6yA_ |
|
not modelled |
9.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
|
|
|
| 80 | c2pmzL_ |
|
not modelled |
9.4 |
10 |
PDB header:translation, transferase
Chain: L: PDB Molecule:dna-directed rna polymerase subunit l;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
|
|
| 81 | d1bbha_ |
|
not modelled |
9.3 |
13 |
Fold:Four-helical up-and-down bundle
Superfamily:Cytochromes
Family:Cytochrome c'-like |
|
|
| 82 | d2j49a1 |
|
not modelled |
9.3 |
14 |
Fold:Taf5 N-terminal domain-like
Superfamily:Taf5 N-terminal domain-like
Family:Taf5 N-terminal domain-like |
|
|
| 83 | c2ld7B_ |
|
not modelled |
9.3 |
8 |
PDB header:transcription
Chain: B: PDB Molecule:paired amphipathic helix protein sin3a;
PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
|
|
|
| 84 | c4o9lA_ |
|
not modelled |
9.2 |
22 |
PDB header:antiviral protein
Chain: A: PDB Molecule:mitochondrial antiviral signaling protein (mavs);
PDBTitle: crystal structure of horse mavs card domain mutant e26r
|
|
|
| 85 | c2vyaB_ |
|
not modelled |
9.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:fatty-acid amide hydrolase 1;
PDBTitle: crystal structure of fatty acid amide hydrolase conjugated2 with the drug-like inhibitor pf-750
|
|
|
| 86 | c3kfuE_ |
|
not modelled |
9.1 |
15 |
PDB header:ligase/rna
Chain: E: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit a;
PDBTitle: crystal structure of the transamidosome
|
|
|
| 87 | c2k42A_ |
|
not modelled |
9.0 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:wiskott-aldrich syndrome protein;
PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
|
|
|
| 88 | d2pv7a1 |
|
not modelled |
9.0 |
8 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:TyrA dimerization domain-like |
|
|
| 89 | d1pswa_ |
|
not modelled |
9.0 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:ADP-heptose LPS heptosyltransferase II |
|
|
| 90 | d2nxpa1 |
|
not modelled |
8.9 |
31 |
Fold:Taf5 N-terminal domain-like
Superfamily:Taf5 N-terminal domain-like
Family:Taf5 N-terminal domain-like |
|
|
| 91 | d1zyma1 |
|
not modelled |
8.5 |
11 |
Fold:SAM domain-like
Superfamily:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain
Family:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain |
|
|
| 92 | c3o59X_ |
|
not modelled |
8.4 |
22 |
PDB header:transferase
Chain: X: PDB Molecule:dna polymerase ii large subunit;
PDBTitle: dna polymerase d large subunit dp2(1-300) from pyrococcus horikoshii
|
|
|
| 93 | c6mn5A_ |
|
not modelled |
8.3 |
24 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:aminoglycoside n(3)-acetyltransferase, aac(3)-iva;
PDBTitle: crystal structure of aminoglycoside acetyltransferase aac(3)-iva,2 h154a mutant, in complex with gentamicin c1a
|
|
|
| 94 | c4dhxC_ |
|
not modelled |
8.3 |
19 |
PDB header:transport protein/dna binding protein
Chain: C: PDB Molecule:enhancer of yellow 2 transcription factor homolog;
PDBTitle: eny2:ganp complex
|
|
|
| 95 | d3bzka2 |
|
not modelled |
8.3 |
21 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Tex HhH-containing domain-like |
|
|
| 96 | c4c2mK_ |
|
not modelled |
8.2 |
17 |
PDB header:transcription
Chain: K: PDB Molecule:dna-directed rna polymerases i and iii subunit rpac2;
PDBTitle: structure of rna polymerase i at 2.8 a resolution
|
|
|
| 97 | c6hqgB_ |
|
not modelled |
8.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450-153 from phenylobacterium zucineum
|
|
|
| 98 | c3fnnA_ |
|
not modelled |
7.9 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:thymidylate synthase thyx;
PDBTitle: biochemical and structural analysis of an atypical thyx:2 corynebacterium glutamicum nchu 87078 depends on thya for3 thymidine biosynthesis
|
|
|
| 99 | d1omha_ |
|
not modelled |
7.9 |
16 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Relaxase domain |
|
|