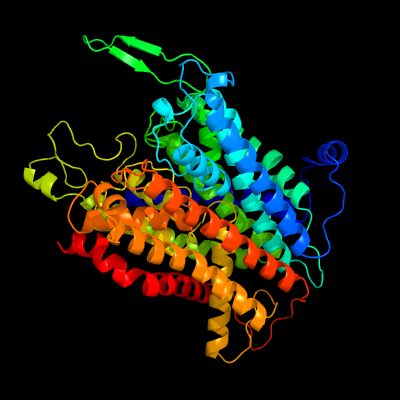
| 1 | c5mrwA_
|
|
 |
100.0 |
51 |
PDB header:hydrolase
Chain: A: PDB Molecule:potassium-transporting atpase potassium-binding subunit;
PDBTitle: structure of the kdpfabc complex
|
|
|
|
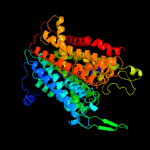
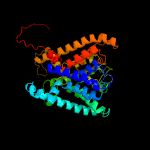
| 2 | c4j7cK_
|
|
 |
100.0 |
15 |
PDB header:transport protein
Chain: K: PDB Molecule:ktr system potassium uptake protein b;
PDBTitle: ktrab potassium transporter from bacillus subtilis
|
|
|
|
| 3 | c3pjzB_
|
|
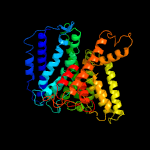 |
99.7 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:potassium uptake protein trkh;
PDBTitle: crystal structure of the potassium transporter trkh from vibrio2 parahaemolyticus
|
|
|
|
| 4 | c3pjzA_
|
|
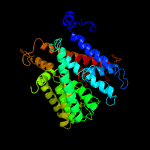 |
99.5 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:potassium uptake protein trkh;
PDBTitle: crystal structure of the potassium transporter trkh from vibrio2 parahaemolyticus
|
|
|
|
| 5 | c6o7ua_
|
|
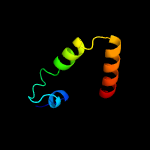 |
52.4 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:
PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
|
|
|
|
| 6 | c2ndjA_
|
|
 |
39.1 |
35 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e member 3;
PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
|
|
|
|
| 7 | c4ev6E_
|
|
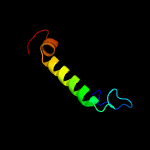 |
27.9 |
24 |
PDB header:metal transport
Chain: E: PDB Molecule:magnesium transport protein cora;
PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
|
|
|
|
| 8 | d1wmxa_
|
|
 |
24.8 |
38 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 30 carbohydrate binding module, CBM30 (PKD repeat) |
|
|
|
| 9 | c6o7xa_
|
|
 |
22.0 |
11 |
PDB header:membrane protein
Chain: A: PDB Molecule:vacuolar atp synthase catalytic subunit a;
PDBTitle: saccharomyces cerevisiae v-atpase stv1-v1vo state 3
|
|
|
|
| 10 | d1wmxb_
|
|
 |
21.6 |
43 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 30 carbohydrate binding module, CBM30 (PKD repeat) |
|
|
|
| 11 | c5nhuI_
|
|
 |
16.0 |
71 |
PDB header:hydrolase
Chain: I: PDB Molecule:agap008004-pa;
PDBTitle: human alpha thrombin complexed with anopheles gambiae ce52 anticoagulant
|
|
|
|
| 12 | c6cfwF_
|
|
 |
15.6 |
25 |
PDB header:membrane protein
Chain: F: PDB Molecule:monovalent cation/h+ antiporter subunit b;
PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
|
|
|
|
| 13 | c3ogiC_
|
|
 |
15.5 |
60 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative esat-6-like protein 6;
PDBTitle: crystal structure of the mycobacterium tuberculosis h37rv esxop2 complex (rv2346c-rv2347c)
|
|
|
|
| 14 | c4gzrA_
|
|
 |
15.2 |
60 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:esat-6-like protein 6;
PDBTitle: crystal structure of the mycobacterium tuberculosis h37rv esxop2 (rv2346c-rv2347c) complex in space group c2221
|
|
|
|
| 15 | c2k21A_
|
|
 |
15.1 |
27 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
|
|
|
|
| 16 | c3j47O_
|
|
 |
14.2 |
60 |
PDB header:protein binding
Chain: O: PDB Molecule:26s proteasome regulatory subunit rpn9;
PDBTitle: formation of an intricate helical bundle dictates the assembly of the2 26s proteasome lid
|
|
|
|
| 17 | c5nhuJ_
|
|
 |
12.7 |
71 |
PDB header:hydrolase
Chain: J: PDB Molecule:agap008004-pa;
PDBTitle: human alpha thrombin complexed with anopheles gambiae ce52 anticoagulant
|
|
|
|
| 18 | c5n9yB_
|
|
 |
12.5 |
20 |
PDB header:membrane protein
Chain: B: PDB Molecule:zinc transport protein zntb;
PDBTitle: the full-length structure of zntb
|
|
|
|
| 19 | d1oe8a1
|
|
 |
11.9 |
13 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
|
|
|
| 20 | d1gu3a_
|
|
 |
11.5 |
8 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:CBM4/9 |
|
|
|
| 21 | c5yf4A_ |
|
not modelled |
11.3 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:mob-like protein phocein;
PDBTitle: a kinase complex mst4-mob4
|
|
|
| 22 | c5halA_ |
|
not modelled |
11.2 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative beta-lactamase from burkholderia2 vietnamiensis
|
|
|
| 23 | c1ot0A_ |
|
not modelled |
11.0 |
25 |
PDB header:antibiotic
Chain: A: PDB Molecule:50s ribosomal protein l1;
PDBTitle: structure of antimicrobial peptide, hp (2-20) and its2 analogues derived from helicobacter pylori, as determined3 by 1h nmr spectroscopy
|
|
|
| 24 | c1p0jA_ |
|
not modelled |
11.0 |
25 |
PDB header:ribosome
Chain: A: PDB Molecule:19-mer peptide from 50s ribosomal protein l1;
PDBTitle: hp (2-20) substitution asp to trp modification in sds-d252 micelles
|
|
|
| 25 | c5wylB_ |
|
not modelled |
11.0 |
40 |
PDB header:ribosomal protein/nuclear protein
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of chaetomium thermophilum utp10 n-terminal domain2 in complex with utp17 c-terminal helices
|
|
|
| 26 | c2kk2A_ |
|
not modelled |
10.9 |
38 |
PDB header:signaling protein
Chain: A: PDB Molecule:en-a1;
PDBTitle: nmr solution structure of the pheromone en-a1 from euplotes nobilii
|
|
|
| 27 | d1d02a_ |
|
not modelled |
10.7 |
57 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease MunI |
|
|
| 28 | c5twvG_ |
|
not modelled |
10.7 |
15 |
PDB header:transport protein
Chain: G: PDB Molecule:atp-sensitive inward rectifier potassium channel 11;
PDBTitle: cryo-em structure of the pancreatic atp-sensitive k+ channel2 sur1/kir6.2 in the presence of atp and glibenclamide
|
|
|
| 29 | c5wylD_ |
|
not modelled |
10.6 |
40 |
PDB header:ribosomal protein/nuclear protein
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of chaetomium thermophilum utp10 n-terminal domain2 in complex with utp17 c-terminal helices
|
|
|
| 30 | c2v52M_ |
|
not modelled |
10.3 |
38 |
PDB header:structural protein/contractile protein
Chain: M: PDB Molecule:mkl/myocardin-like protein 1;
PDBTitle: structure of mal-rpel2 complexed to g-actin
|
|
|
| 31 | c2levA_ |
|
not modelled |
10.2 |
53 |
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:ler;
PDBTitle: structure of the dna complex of the c-terminal domain of ler
|
|
|
| 32 | c1zcdA_ |
|
not modelled |
10.1 |
21 |
PDB header:membrane protein
Chain: A: PDB Molecule:na(+)/h(+) antiporter 1;
PDBTitle: crystal structure of the na+/h+ antiporter nhaa
|
|
|
| 33 | c3lw5K_ |
|
not modelled |
9.6 |
31 |
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit x psak;
PDBTitle: improved model of plant photosystem i
|
|
|
| 34 | c2bpqB_ |
|
not modelled |
9.5 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase (trpd) from2 mycobacterium tuberculosis (apo structure)
|
|
|
| 35 | d1kz1a_ |
|
not modelled |
9.5 |
18 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
|
|
| 36 | c2m0qA_ |
|
not modelled |
9.5 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e member 2;
PDBTitle: solution nmr analysis of intact kcne2 in detergent micelles2 demonstrate a straight transmembrane helix
|
|
|
| 37 | d1hnra_ |
|
not modelled |
9.4 |
41 |
Fold:H-NS histone-like proteins
Superfamily:H-NS histone-like proteins
Family:H-NS histone-like proteins |
|
|
| 38 | c5xyvC_ |
|
not modelled |
9.4 |
26 |
PDB header:protein binding
Chain: C: PDB Molecule:protein deadlock;
PDBTitle: crystal structure of drosophila melanogaster rhino chromoshadow domain2 in complex with deadlock n-terminal domain
|
|
|
| 39 | c4p22A_ |
|
not modelled |
9.2 |
31 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme 1;
PDBTitle: crystal structure of n-terminal fragments of e1
|
|
|
| 40 | c2wscK_ |
|
not modelled |
9.0 |
38 |
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit psak, chloroplastic;
PDBTitle: improved model of plant photosystem i
|
|
|
| 41 | c5zghJ_ |
|
not modelled |
8.9 |
32 |
PDB header:photosynthesis
Chain: J: PDB Molecule:psaj;
PDBTitle: cryo-em structure of the red algal psi-lhcr
|
|
|
| 42 | c3nrlB_ |
|
not modelled |
8.6 |
36 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein rumgna_01417;
PDBTitle: crystal structure of protein rumgna_01417 from ruminococcus gnavus,2 northeast structural genomics consortium target ugr76
|
|
|
| 43 | d1gvha3 |
|
not modelled |
8.6 |
24 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Flavohemoglobin, C-terminal domain |
|
|
| 44 | d1nvmb1 |
|
not modelled |
8.5 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 45 | d1yova1 |
|
not modelled |
8.3 |
38 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Ubiquitin activating enzymes (UBA) |
|
|
| 46 | d1d4oa_ |
|
not modelled |
8.1 |
67 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
|
|
| 47 | c1pt9B_ |
|
not modelled |
8.1 |
67 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase, mitochondrial;
PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
|
|
|
| 48 | d1jb0j_ |
|
not modelled |
8.0 |
21 |
Fold:Single transmembrane helix
Superfamily:Subunit IX of photosystem I reaction centre, PsaJ
Family:Subunit IX of photosystem I reaction centre, PsaJ |
|
|
| 49 | c2ks1A_ |
|
not modelled |
8.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
|
|
|
| 50 | c2jwaA_ |
|
not modelled |
8.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
|
|
|
| 51 | d2cnda2 |
|
not modelled |
7.9 |
10 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
|
|
| 52 | c3cmmA_ |
|
not modelled |
7.9 |
23 |
PDB header:ligase/protein binding
Chain: A: PDB Molecule:ubiquitin-activating enzyme e1 1;
PDBTitle: crystal structure of the uba1-ubiquitin complex
|
|
|
| 53 | d1pnoa_ |
|
not modelled |
7.8 |
44 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
|
|
| 54 | c6igzK_ |
|
not modelled |
7.8 |
36 |
PDB header:plant protein
Chain: K: PDB Molecule:psak;
PDBTitle: structure of psi-lhci
|
|
|
| 55 | c6dc6A_ |
|
not modelled |
7.8 |
31 |
PDB header:signaling protein/ligase
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme 1;
PDBTitle: crystal structure of human ubiquitin activating enzyme e1 (uba1) in2 complex with ubiquitin
|
|
|
| 56 | c4ky0B_ |
|
not modelled |
7.7 |
18 |
PDB header:transport protein, membrane protein
Chain: B: PDB Molecule:proton/glutamate symporter, sdf family;
PDBTitle: crystal structure of a substrate-free glutamate transporter homologue2 from thermococcus kodakarensis
|
|
|
| 57 | c2o01J_ |
|
not modelled |
7.5 |
32 |
PDB header:photosynthesis
Chain: J: PDB Molecule:photosystem i reaction center subunit ix;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.4 angstrom2 resolution
|
|
|
| 58 | c5xtrC_ |
|
not modelled |
7.4 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:fad-linked sulfhydryl oxidase;
PDBTitle: crystal structure of baculoviral sulfhydryl oxidase p33 (r127a, e183a2 mutant)
|
|
|
| 59 | c3j9pD_ |
|
not modelled |
7.3 |
27 |
PDB header:transport protein
Chain: D: PDB Molecule:maltose-binding periplasmic protein, transient receptor
PDBTitle: structure of the trpa1 ion channel determined by electron cryo-2 microscopy
|
|
|
| 60 | c6rdi8_ |
|
not modelled |
7.2 |
42 |
PDB header:proton transport
Chain: 8: PDB Molecule:mitochondrial atp synthase subunit asa8;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1a,2 monomer-masked refinement
|
|
|
| 61 | c6rdr8_ |
|
not modelled |
7.2 |
42 |
PDB header:proton transport
Chain: 8: PDB Molecule:mitochondrial atp synthase subunit asa8;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1d,2 monomer-masked refinement
|
|
|
| 62 | c2bruC_ |
|
not modelled |
7.1 |
56 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad(p) transhydrogenase subunit beta;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
|
|
| 63 | c4gtnA_ |
|
not modelled |
7.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: structure of anthranilate phosphoribosyl transferase from2 acinetobacter baylyi
|
|
|
| 64 | d1g3wa2 |
|
not modelled |
7.1 |
19 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
|
|
| 65 | c5h5pA_ |
|
not modelled |
7.0 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:myelin regulatory factor;
PDBTitle: crystal structure of myelin-gene regulatory factor dna binding domain
|
|
|
| 66 | c4ii3A_ |
|
not modelled |
7.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-activating enzyme e1 1;
PDBTitle: crystal structure of s. pombe ubiquitin activating enzyme 1 (uba1) in2 complex with ubiquitin and atp/mg
|
|
|
| 67 | c2yf3F_ |
|
not modelled |
6.9 |
33 |
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from deinococcus2 radiodurans, complex with manganese
|
|
|
| 68 | c2yn9B_ |
|
not modelled |
6.7 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: cryo-em structure of gastric h+,k+-atpase with bound rubidium
|
|
|
| 69 | d2h8pc1 |
|
not modelled |
6.6 |
30 |
Fold:Voltage-gated potassium channels
Superfamily:Voltage-gated potassium channels
Family:Voltage-gated potassium channels |
|
|
| 70 | c6ijjK_ |
|
not modelled |
6.4 |
17 |
PDB header:membrane protein
Chain: K: PDB Molecule:psak;
PDBTitle: photosystem i of chlamydomonas reinhardtii
|
|
|
| 71 | c3dxeB_ |
|
not modelled |
6.3 |
43 |
PDB header:protein binding
Chain: B: PDB Molecule:amyloid beta a4 protein;
PDBTitle: crystal structure of the intracellular domain of human app (t668a2 mutant) in complex with fe65-ptb2
|
|
|
| 72 | d1tvca2 |
|
not modelled |
6.2 |
20 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Aromatic dioxygenase reductase-like |
|
|
| 73 | d2piaa2 |
|
not modelled |
6.2 |
15 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Aromatic dioxygenase reductase-like |
|
|
| 74 | c5mqfT_ |
|
not modelled |
6.2 |
19 |
PDB header:splicing
Chain: T: PDB Molecule:pre-mrna-splicing factor cwc22 homolog;
PDBTitle: cryo-em structure of a human spliceosome activated for step 2 of2 splicing (c* complex)
|
|
|
| 75 | c6ijoG_ |
|
not modelled |
6.1 |
33 |
PDB header:photosynthesis
Chain: G: PDB Molecule:psag;
PDBTitle: photosystem i of chlamydomonas reinhardtii
|
|
|
| 76 | c3qnqD_ |
|
not modelled |
6.0 |
15 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
|
|
| 77 | d2c7na1 |
|
not modelled |
6.0 |
30 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:A20-like zinc finger |
|
|
| 78 | d1usha1 |
|
not modelled |
5.7 |
17 |
Fold:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
Superfamily:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain |
|
|
| 79 | d2isya2 |
|
not modelled |
5.7 |
21 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
|
|
| 80 | c2zxeB_ |
|
not modelled |
5.7 |
17 |
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:na+,k+-atpase beta subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
|
|
|
| 81 | c1nvmB_ |
|
not modelled |
5.7 |
32 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase (acylating);
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
| 82 | c2epgB_ |
|
not modelled |
5.6 |
40 |
PDB header:ligase
Chain: B: PDB Molecule:hypothetical protein ttha1785;
PDBTitle: crystal structure of ttha1785
|
|
|
| 83 | d1qfja2 |
|
not modelled |
5.6 |
4 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
|
|
| 84 | c4ainB_ |
|
not modelled |
5.6 |
16 |
PDB header:membrane protein
Chain: B: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of betp with asymmetric protomers.
|
|
|
| 85 | c2lsjA_ |
|
not modelled |
5.5 |
25 |
PDB header:protein binding/protein binding
Chain: A: PDB Molecule:dna repair protein rev1;
PDBTitle: solution structure of the mouse rev1 ctd in complex with the rev1-2 interacting region (rir)of pol kappa
|
|
|
| 86 | c5z47A_ |
|
not modelled |
5.5 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of pyrrolidone carboxylate peptidase i with2 disordered loop a from deinococcus radiodurans r1
|
|
|
| 87 | c2ot8D_ |
|
not modelled |
5.5 |
33 |
PDB header:transport protein
Chain: D: PDB Molecule:heterogeneous nuclear ribonucleoprotein m;
PDBTitle: karyopherin beta2/transportin-hnrnpm nls complex
|
|
|
| 88 | c4nkpD_ |
|
not modelled |
5.4 |
18 |
PDB header:chaperone
Chain: D: PDB Molecule:putative extracellular heme-binding protein;
PDBTitle: crystal structure of a putative extracellular heme-binding protein2 (despig_02683) from desulfovibrio piger atcc 29098 at 1.24 a3 resolution
|
|
|
| 89 | d1fdra2 |
|
not modelled |
5.3 |
4 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
|
|
| 90 | c4uecB_ |
|
not modelled |
5.3 |
36 |
PDB header:translation
Chain: B: PDB Molecule:eukaryotic translation initiation factor 4g, isoform a;
PDBTitle: complex of d. melanogaster eif4e with eif4g and cap analog
|
|
|
| 91 | d1x7fa1 |
|
not modelled |
5.3 |
40 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:Outer surface protein, C-terminal domain |
|
|
| 92 | c3mk7F_ |
|
not modelled |
5.2 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
|
|
|
| 93 | c2c7mA_ |
|
not modelled |
5.2 |
30 |
PDB header:protein binding
Chain: A: PDB Molecule:rab guanine nucleotide exchange factor 1;
PDBTitle: human rabex-5 residues 1-74 in complex with ubiquitin
|
|
|
| 94 | d1krha2 |
|
not modelled |
5.2 |
12 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Aromatic dioxygenase reductase-like |
|
|