| 1 | c5mrwG_
|
|
 |
100.0 |
46 |
PDB header:hydrolase
Chain: G: PDB Molecule:potassium-transporting atpase kdpc subunit;
PDBTitle: structure of the kdpfabc complex
|
|
|
|
| 2 | c4oxqB_
|
|
 |
73.5 |
17 |
PDB header:metal binding protein
Chain: B: PDB Molecule:manganese abc transporter, periplasmic-binding protein
PDBTitle: structure of staphylococcus pseudintermedius metal-binding protein2 sita in complex with zinc
|
|
|
|
| 3 | c4k3vA_
|
|
 |
58.7 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:abc superfamily atp binding cassette transporter, binding
PDBTitle: structure of staphylococcus aureus mntc
|
|
|
|
| 4 | d1hq1a_
|
|
 |
55.4 |
25 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
|
|
|
| 5 | d1dula_
|
|
 |
54.5 |
23 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
|
|
|
| 6 | d1qb2a_
|
|
 |
53.3 |
33 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
|
|
|
| 7 | c4aktB_
|
|
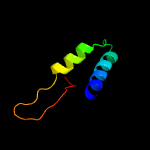 |
53.1 |
22 |
PDB header:hydrolase/peptide
Chain: B: PDB Molecule:thiazoline oxidase/subtilisin-like protease;
PDBTitle: patg macrocyclase in complex with peptide
|
|
|
|
| 8 | c4ue4C_
|
|
 |
51.8 |
27 |
PDB header:translation
Chain: C: PDB Molecule:signal recognition particle protein;
PDBTitle: structural basis for targeting and elongation arrest of bacillus2 signal recognition particle
|
|
|
|
| 9 | c2jqeA_
|
|
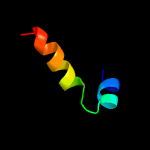 |
51.4 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: soution structure of af54 m-domain
|
|
|
|
| 10 | c4xcoC_
|
|
 |
50.0 |
12 |
PDB header:rna binding protein
Chain: C: PDB Molecule:signal recognition particle 54 kda protein,signal sequence;
PDBTitle: signal-sequence induced conformational changes in the signal2 recognition particle
|
|
|
|
| 11 | d2ffha2
|
|
 |
49.1 |
27 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
|
|
|
| 12 | c3ipwA_
|
|
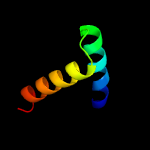 |
47.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase tatd family protein;
PDBTitle: crystal structure of hydrolase tatd family protein from entamoeba2 histolytica
|
|
|
|
| 13 | d1qzxa2
|
|
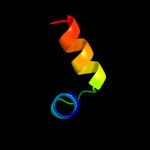 |
46.2 |
23 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
|
|
|
| 14 | c3e2vA_
|
|
 |
43.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:3'-5'-exonuclease;
PDBTitle: crystal structure of an uncharacterized amidohydrolase from2 saccharomyces cerevisiae
|
|
|
|
| 15 | c3rcmA_
|
|
 |
43.5 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:tatd family hydrolase;
PDBTitle: crystal structure of efi target 500140:tatd family hydrolase from2 pseudomonas putida
|
|
|
|
| 16 | c3gg7A_
|
|
 |
41.2 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized metalloprotein;
PDBTitle: crystal structure of an uncharacterized metalloprotein from2 deinococcus radiodurans
|
|
|
|
| 17 | d1xwya1
|
|
 |
35.6 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
|
|
|
| 18 | d1j6oa_
|
|
 |
35.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
|
|
|
| 19 | d1zzma1
|
|
 |
28.3 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
|
|
|
| 20 | c2y1hA_
|
|
 |
26.1 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deoxyribonuclease tatdn3;
PDBTitle: crystal structure of the human tatd-domain protein 3 (tatdn3)
|
|
|
|
| 21 | c3qllB_ |
|
not modelled |
25.0 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase;
PDBTitle: crystal structure of ripc from yersinia pestis
|
|
|
| 22 | c4cl2A_ |
|
not modelled |
23.4 |
29 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic solute binding protein;
PDBTitle: structure of periplasmic metal binding protein from candidatus2 liberibacter asiaticus
|
|
|
| 23 | c1toaA_ |
|
not modelled |
23.2 |
17 |
PDB header:binding protein
Chain: A: PDB Molecule:protein (periplasmic binding protein troa);
PDBTitle: periplasmic zinc binding protein troa from treponema pallidum
|
|
|
| 24 | d1toaa_ |
|
not modelled |
23.2 |
17 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
|
|
| 25 | c2xioA_ |
|
not modelled |
22.7 |
6 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deoxyribonuclease tatdn1;
PDBTitle: structure of putative deoxyribonuclease tatdn1 isoform a
|
|
|
| 26 | c4ysnD_ |
|
not modelled |
22.5 |
41 |
PDB header:isomerase
Chain: D: PDB Molecule:putative 4-aminobutyrate aminotransferase;
PDBTitle: structure of aminoacid racemase in complex with plp
|
|
|
| 27 | d2ijob1 |
|
not modelled |
21.6 |
33 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
|
|
| 28 | c3e90B_ |
|
not modelled |
21.0 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:ns3 protease;
PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
|
|
|
| 29 | c2ps3A_ |
|
not modelled |
21.0 |
17 |
PDB header:metal transport
Chain: A: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: structure and metal binding properties of znua, a periplasmic zinc2 transporter from escherichia coli
|
|
|
| 30 | d1befa_ |
|
not modelled |
19.9 |
28 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
|
|
| 31 | c2ordA_ |
|
not modelled |
19.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
|
|
|
| 32 | c3lkwA_ |
|
not modelled |
19.8 |
28 |
PDB header:viral protein,hydrolase
Chain: A: PDB Molecule:fusion protein of nonstructural protein 2b and
PDBTitle: crystal structure of dengue virus 1 ns2b/ns3 protease active site2 mutant
|
|
|
| 33 | c2j37W_ |
|
not modelled |
19.2 |
33 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein (srp54);
PDBTitle: model of mammalian srp bound to 80s rncs
|
|
|
| 34 | c3mfqB_ |
|
not modelled |
19.1 |
19 |
PDB header:metal binding protein
Chain: B: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: a glance into the metal binding specificity of troa: where elaborate2 behaviors occur in the active center
|
|
|
| 35 | c5nlaA_ |
|
not modelled |
18.8 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator transcription regulator
PDBTitle: crystal structure of the arac-like transcriptional activator cuxr
|
|
|
| 36 | d2phcb1 |
|
not modelled |
18.6 |
21 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:PH0987 C-terminal domain-like |
|
|
| 37 | c4bjqB_ |
|
not modelled |
18.6 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:penicillin binding protein transpeptidase domain protein;
PDBTitle: crystal structure of e. coli penicillin binding protein 3,2 domain v88-s165
|
|
|
| 38 | d2fomb1 |
|
not modelled |
18.5 |
28 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
|
|
| 39 | c2v3cC_ |
|
not modelled |
18.4 |
13 |
PDB header:signaling protein
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
|
|
|
| 40 | c5lc0B_ |
|
not modelled |
18.3 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:ns2b-ns3 protease,ns2b-ns3 protease;
PDBTitle: crystal structure of zika virus ns2b-ns3 protease in complex with a2 boronate inhibitor
|
|
|
| 41 | c4m9mA_ |
|
not modelled |
18.2 |
28 |
PDB header:viral protein
Chain: A: PDB Molecule:ns2b-ns3 protease;
PDBTitle: ns2b-ns3 protease from dengue virus at ph 8.5
|
|
|
| 42 | d1vkoa1 |
|
not modelled |
18.2 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 43 | c2gzxB_ |
|
not modelled |
18.0 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative tatd related dnase;
PDBTitle: crystal structure of the tatd deoxyribonuclease mw0446 from2 staphylococcus aureus. northeast structural genomics consortium3 target zr237.
|
|
|
| 44 | c1gw4A_ |
|
not modelled |
17.6 |
22 |
PDB header:high density lipoproteins
Chain: A: PDB Molecule:apoa-i;
PDBTitle: the helix-hinge-helix structural motif in human2 apolipoprotein a-i determined by nmr spectroscopy, 13 structure
|
|
|
| 45 | c2yolA_ |
|
not modelled |
17.5 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:serine protease subunit ns2b, serine protease ns3;
PDBTitle: west nile virus ns2b-ns3 protease in complex with 3,4-2 dichlorophenylacetyl-lys-lys-gcma
|
|
|
| 46 | c3dxvA_ |
|
not modelled |
17.0 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:alpha-amino-epsilon-caprolactam racemase;
PDBTitle: the crystal structure of alpha-amino-epsilon-caprolactam racemase from2 achromobacter obae
|
|
|
| 47 | c3dm5A_ |
|
not modelled |
16.7 |
21 |
PDB header:rna binding protein, transport protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: structures of srp54 and srp19, the two proteins assembling the2 ribonucleic core of the signal recognition particle from the archaeon3 pyrococcus furiosus.
|
|
|
| 48 | c5gafi_ |
|
not modelled |
16.6 |
23 |
PDB header:ribosome
Chain: I: PDB Molecule:50s ribosomal protein l10;
PDBTitle: rnc in complex with srp
|
|
|
| 49 | c2iy3A_ |
|
not modelled |
16.5 |
21 |
PDB header:rna-binding
Chain: A: PDB Molecule:signal recognition particle protein,signal recognition
PDBTitle: structure of the e. coli signal regognition particle
|
|
|
| 50 | c6arbA_ |
|
not modelled |
15.2 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:citrate lyase subunit beta-like protein;
PDBTitle: crystal structure of protein cite from mycobacterium tuberculosis in2 complex with magnesium, pyruvate and coenzyme a
|
|
|
| 51 | c4xrvB_ |
|
not modelled |
15.1 |
36 |
PDB header:metal binding protein
Chain: B: PDB Molecule:periplasmic solute binding protein;
PDBTitle: structure of a zn abc transporter substrate binding protein from2 paracoccus denitrificans
|
|
|
| 52 | c3zxxA_ |
|
not modelled |
14.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:subtilisin-like protein;
PDBTitle: structure of self-cleaved protease domain of pata
|
|
|
| 53 | c1qzwC_ |
|
not modelled |
14.6 |
23 |
PDB header:signaling protein/rna
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
|
|
|
| 54 | c2j289_ |
|
not modelled |
14.5 |
23 |
PDB header:ribosome
Chain: 9: PDB Molecule:signal recognition particle 54;
PDBTitle: model of e. coli srp bound to 70s rncs
|
|
|
| 55 | c2zp2B_ |
|
not modelled |
14.5 |
17 |
PDB header:transferase inhibitor
Chain: B: PDB Molecule:kinase a inhibitor;
PDBTitle: c-terminal domain of kipi from bacillus subtilis
|
|
|
| 56 | c5oqmk_ |
|
not modelled |
14.3 |
14 |
PDB header:transcription
Chain: K: PDB Molecule:dna-directed rna polymerase ii subunit rpb11;
PDBTitle: structure of yeast transcription pre-initiation complex with tfiih and2 core mediator
|
|
|
| 57 | c1vkoA_ |
|
not modelled |
14.2 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:inositol-3-phosphate synthase;
PDBTitle: crystal structure of inositol-3-phosphate synthase (ce21227) from2 caenorhabditis elegans at 2.30 a resolution
|
|
|
| 58 | c1x3wB_ |
|
not modelled |
14.1 |
5 |
PDB header:hydrolase
Chain: B: PDB Molecule:uv excision repair protein rad23;
PDBTitle: structure of a peptide:n-glycanase-rad23 complex
|
|
|
| 59 | d1yixa1 |
|
not modelled |
14.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
|
|
| 60 | c5x3tA_ |
|
not modelled |
14.1 |
33 |
PDB header:antitoxin/toxin
Chain: A: PDB Molecule:antitoxin vapb26;
PDBTitle: vapbc from mycobacterium tuberculosis
|
|
|
| 61 | d2fp7b1 |
|
not modelled |
14.0 |
23 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Viral proteases |
|
|
| 62 | c6n39A_ |
|
not modelled |
13.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:dephospho-coa kinase;
PDBTitle: crystal structure of an dephospho-coa kinase coae from mycobacterium2 paratuberculosis
|
|
|
| 63 | d2pv4a1 |
|
not modelled |
13.9 |
32 |
Fold:Sama2622-like
Superfamily:Sama2622-like
Family:Sama2622-like |
|
|
| 64 | c2pb2B_ |
|
not modelled |
13.8 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:acetylornithine/succinyldiaminopimelate aminotransferase;
PDBTitle: structure of biosynthetic n-acetylornithine aminotransferase from2 salmonella typhimurium: studies on substrate specificity and3 inhibitor binding
|
|
|
| 65 | c4aksA_ |
|
not modelled |
13.3 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:thiazoline oxidase/subtilisin-like protease;
PDBTitle: patg macrocyclase domain
|
|
|
| 66 | c5vxsF_ |
|
not modelled |
13.2 |
21 |
PDB header:lyase
Chain: F: PDB Molecule:citrate lyase subunit beta-like protein, mitochondrial;
PDBTitle: crystal structure analysis of human clybl in apo form
|
|
|
| 67 | c3hjtB_ |
|
not modelled |
13.1 |
25 |
PDB header:cell adhesion, transport protein
Chain: B: PDB Molecule:lmb;
PDBTitle: structure of laminin binding protein (lmb) of streptococcus agalactiae2 a bifunctional protein with adhesin and metal transporting activity
|
|
|
| 68 | d1ohwa_ |
|
not modelled |
13.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 69 | c4cdiC_ |
|
not modelled |
13.0 |
21 |
PDB header:membrane protein
Chain: C: PDB Molecule:predicted protein;
PDBTitle: crystal structure of acrb-acrz complex
|
|
|
| 70 | d1oqya3 |
|
not modelled |
12.9 |
23 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
|
|
| 71 | c2fvmA_ |
|
not modelled |
12.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:dihydropyrimidinase;
PDBTitle: crystal structure of dihydropyrimidinase from saccharomyces kluyveri2 in complex with the reaction product n-carbamyl-beta-alanine
|
|
|
| 72 | c2ma3A_ |
|
not modelled |
12.5 |
29 |
PDB header:replication
Chain: A: PDB Molecule:dna replication initiator (cdc21/cdc54);
PDBTitle: nmr solution structure of the c-terminus of the minichromosome2 maintenance protein mcm from methanothermobacter thermautotrophicus
|
|
|
| 73 | c4v1ag_ |
|
not modelled |
12.4 |
23 |
PDB header:ribosome
Chain: G: PDB Molecule:
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
|
|
|
| 74 | c6gcsf_ |
|
not modelled |
11.3 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:nufm subunit;
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
| 75 | c5n9jB_ |
|
not modelled |
11.3 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:mediator of rna polymerase ii transcription subunit 10;
PDBTitle: core mediator of transcriptional regulation
|
|
|
| 76 | d2r6gf1 |
|
not modelled |
11.1 |
21 |
Fold:MalF N-terminal region-like
Superfamily:MalF N-terminal region-like
Family:MalF N-terminal region-like |
|
|
| 77 | c1wnfA_ |
|
not modelled |
11.1 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of ph0066 from pyrococcus horikoshii
|
|
|
| 78 | c2ykyB_ |
|
not modelled |
11.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:beta-transaminase;
PDBTitle: structural determinants of the beta-selectivity of a bacterial2 aminotransferase
|
|
|
| 79 | d1e5pa_ |
|
not modelled |
10.6 |
14 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
|
|
| 80 | d1afwa2 |
|
not modelled |
10.6 |
23 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
|
|
| 81 | c3thgA_ |
|
not modelled |
10.6 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase activase 1,
PDBTitle: crystal structure of the creosote rubisco activase c-domain
|
|
|
| 82 | c1p1hD_ |
|
not modelled |
10.5 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:inositol-3-phosphate synthase;
PDBTitle: crystal structure of the 1l-myo-inositol/nad+ complex
|
|
|
| 83 | c3vs9F_ |
|
not modelled |
9.9 |
17 |
PDB header:transferase
Chain: F: PDB Molecule:type iii polyketide synthase;
PDBTitle: crystal structure of type iii pks arsc mutant
|
|
|
| 84 | d1d8ca_ |
|
not modelled |
9.9 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Malate synthase G
Family:Malate synthase G |
|
|
| 85 | c4oddB_ |
|
not modelled |
9.9 |
7 |
PDB header:allergen
Chain: B: PDB Molecule:lipocalin allergen;
PDBTitle: crystal structure of a dog lipocalin allergen
|
|
|
| 86 | c3beyC_ |
|
not modelled |
9.8 |
8 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:conserved protein o27018;
PDBTitle: crystal structure of the protein o27018 from methanobacterium2 thermoautotrophicum. northeast structural genomics consortium target3 tt217
|
|
|
| 87 | c2k9sA_ |
|
not modelled |
9.7 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:arabinose operon regulatory protein;
PDBTitle: solution structure of dna binding domain of e. coli arac
|
|
|
| 88 | c5mmjw_ |
|
not modelled |
9.6 |
57 |
PDB header:ribosome
Chain: W: PDB Molecule:
PDBTitle: structure of the small subunit of the chloroplast ribosome
|
|
|
| 89 | c1u5vA_ |
|
not modelled |
9.6 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:cite;
PDBTitle: structure of cite complexed with triphosphate group of atp form2 mycobacterium tuberculosis
|
|
|
| 90 | d1gnsa_ |
|
not modelled |
9.5 |
21 |
Fold:Subtilisin-like
Superfamily:Subtilisin-like
Family:Subtilases |
|
|
| 91 | c2as9B_ |
|
not modelled |
9.5 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:serine protease;
PDBTitle: functional and structural characterization of spl proteases from2 staphylococcus aureus
|
|
|
| 92 | d1tw3a1 |
|
not modelled |
9.4 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
|
|
| 93 | c2ee7A_ |
|
not modelled |
9.3 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:sperm flagellar protein 1;
PDBTitle: solution structure of the ch domain from human sperm2 flagellar protein 1
|
|
|
| 94 | c2ov3A_ |
|
not modelled |
9.3 |
33 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein component of an abc type zinc
PDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc bound
|
|
|
| 95 | d1rp3a1 |
|
not modelled |
9.2 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
|
|
| 96 | c3c19A_ |
|
not modelled |
9.2 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mk0293;
PDBTitle: crystal structure of protein mk0293 from methanopyrus kandleri av19
|
|
|
| 97 | c2mxbA_ |
|
not modelled |
9.2 |
50 |
PDB header:membrane protein
Chain: A: PDB Molecule:erythropoietin receptor;
PDBTitle: structure of the transmembrane domain of the mouse erythropoietin2 receptor
|
|
|
| 98 | c5xtdo_ |
|
not modelled |
9.1 |
23 |
PDB header:oxidoreductase/electron transport
Chain: O: PDB Molecule:nadh dehydrogenase [ubiquinone] flavoprotein 2,
PDBTitle: cryo-em structure of human respiratory complex i
|
|
|
| 99 | c2huoA_ |
|
not modelled |
9.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol oxygenase;
PDBTitle: crystal structure of mouse myo-inositol oxygenase in complex with2 substrate
|
|
|