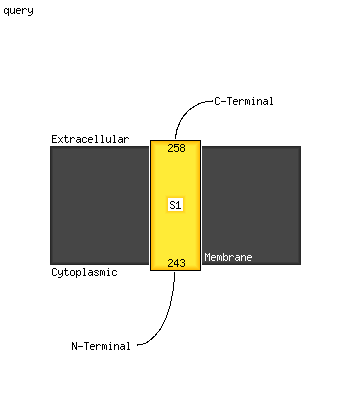
1 c5xfsB_
100.0
100
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 d2g38b1
100.0
31
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
3 c2g38B_
100.0
31
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
4 c4xy3A_
100.0
17
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
98.8
17
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c2vs0B_
97.9
11
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
7 c4iogD_
97.7
10
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
8 c3gvmA_
97.7
11
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
97.5
18
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
96.8
17
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c4lwsB_
95.4
17
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
12 c4lwsA_
94.5
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
13 d1wa8b1
94.4
20
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
14 c4i0xA_
93.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
15 c2kg7B_
88.2
18
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
16 c4i0xJ_
63.0
20
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
17 c5frgA_
17.5
75
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
18 d1ui5a2
17.4
24
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
19 c1bkvA_
16.8
56
PDB header: structural proteinChain: A: PDB Molecule: t3-785;PDBTitle: collagen
20 c1bkvC_
16.0
56
PDB header: structural proteinChain: C: PDB Molecule: t3-785;PDBTitle: collagen
21 c1bkvB_
not modelled
16.0
56
PDB header: structural proteinChain: B: PDB Molecule: t3-785;PDBTitle: collagen
22 c2ke4A_
not modelled
10.3
75
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
23 c1vytF_
not modelled
7.1
50
PDB header: transport proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channelPDBTitle: beta3 subunit complexed with aid
24 c4xb6D_
not modelled
7.0
20
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
25 c4gyxC_
not modelled
6.9
45
PDB header: structural protein, blood clottingChain: C: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
26 c2iu1A_
not modelled
6.7
22
PDB header: transcriptionChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of eif5 c-terminal domain
27 c4dmtC_
not modelled
6.7
38
PDB header: structural proteinChain: C: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
28 c4dmtB_
not modelled
6.7
38
PDB header: structural proteinChain: B: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
29 c4dmtA_
not modelled
6.7
38
PDB header: structural proteinChain: A: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
30 c4gyxA_
not modelled
6.6
45
PDB header: structural protein, blood clottingChain: A: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
31 c4gyxB_
not modelled
6.6
45
PDB header: structural protein, blood clottingChain: B: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
32 d1fcda3
not modelled
6.4
31
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
33 c2fulE_
not modelled
6.1
33
PDB header: translationChain: E: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of the c-terminal domain of s. cerevisiae eif5
34 c2kg7A_
not modelled
6.0
35
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein esxg (pe family protein);PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
35 c1vytE_
not modelled
5.7
50
PDB header: transport proteinChain: E: PDB Molecule: voltage-dependent l-type calcium channelPDBTitle: beta3 subunit complexed with aid
36 c2lkqA_
not modelled
5.6
56
PDB header: immune systemChain: A: PDB Molecule: immunoglobulin lambda-like polypeptide 1;PDBTitle: nmr structure of the lambda 5 22-45 peptide
37 c2y5tG_
not modelled
5.6
67
PDB header: immune systemChain: G: PDB Molecule: c1;PDBTitle: crystal structure of the pathogenic autoantibody ciic1 in complex with2 the triple-helical c1 peptide
38 c6q5lA_
not modelled
5.5
23
PDB header: de novo proteinChain: A: PDB Molecule: cc-hex*-l24h;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24h
39 c6q5lB_
not modelled
5.5
23
PDB header: de novo proteinChain: B: PDB Molecule: cc-hex*-l24h;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24h
40 c6q5hA_
not modelled
5.5
23
PDB header: de novo proteinChain: A: PDB Molecule: cc-hex*-l24d;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24d
41 c6q5mB_
not modelled
5.4
23
PDB header: de novo proteinChain: B: PDB Molecule: cc-hex*-l24dab;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24dab
42 c6q5iB_
not modelled
5.4
23
PDB header: de novo proteinChain: B: PDB Molecule: cc-hex*-l24e;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24e
43 c6q5kA_
not modelled
5.4
23
PDB header: de novo proteinChain: A: PDB Molecule: cc-hex*-l24k;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24k
44 d1zeea1
not modelled
5.4
33
Fold: Indolic compounds 2,3-dioxygenase-likeSuperfamily: Indolic compounds 2,3-dioxygenase-likeFamily: Indoleamine 2,3-dioxygenase-like
45 c4lzxB_
not modelled
5.3
38
PDB header: metal binding proteinChain: B: PDB Molecule: iq domain-containing protein g;PDBTitle: complex of iqcg and ca2+-free cam
46 c6q5mA_
not modelled
5.3
23
PDB header: de novo proteinChain: A: PDB Molecule: cc-hex*-l24dab;PDBTitle: crystal structure of a cc-hex mutant that forms an antiparallel four-2 helix coiled coil cc-hex*-l24dab
47 c5uc0B_
not modelled
5.3
60
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein cog5400;PDBTitle: crystal structure of beta-barrel-like, uncharacterized protein of2 cog5400 from brucella abortus
48 c2y5tE_
not modelled
5.3
67
PDB header: immune systemChain: E: PDB Molecule: c1;PDBTitle: crystal structure of the pathogenic autoantibody ciic1 in complex with2 the triple-helical c1 peptide
49 c6q5jE_
not modelled
5.1
23
PDB header: de novo proteinChain: E: PDB Molecule: cc-hex*-l24e;PDBTitle: crystal structure of a cc-hex mutant that forms a parallel six-helix2 coiled coil cc-hex*-l24e
50 c3r47J_
not modelled
5.1
23
PDB header: de novo proteinChain: J: PDB Molecule: coiled coil helix l24h;PDBTitle: crystal structure of a 6-helix coiled coil cc-hex-h24
51 c3r47B_
not modelled
5.1
23
PDB header: de novo proteinChain: B: PDB Molecule: coiled coil helix l24h;PDBTitle: crystal structure of a 6-helix coiled coil cc-hex-h24
52 c3r47I_
not modelled
5.1
23
PDB header: de novo proteinChain: I: PDB Molecule: coiled coil helix l24h;PDBTitle: crystal structure of a 6-helix coiled coil cc-hex-h24
53 c5l85B_
not modelled
5.1
18
PDB header: signaling proteinChain: B: PDB Molecule: nuclear fragile x mental retardation-interacting protein 1;PDBTitle: solution structure of the complex between human znhit3 and nufip12 proteins
54 c4dexB_
not modelled
5.1
38
PDB header: transport proteinChain: B: PDB Molecule: voltage-dependent n-type calcium channel subunit alpha-1b;PDBTitle: crystal structure of the voltage dependent calcium channel beta-22 subunit in complex with the cav2.2 i-ii linker.
55 c6aokA_
not modelled
5.1
38
PDB header: hydrolaseChain: A: PDB Molecule: ceg4;PDBTitle: crystal structure of legionella pneumophila effector ceg4 with n-2 terminal tev protease cleavage sequence
56 c6q5jF_
not modelled
5.1
23
PDB header: de novo proteinChain: F: PDB Molecule: cc-hex*-l24e;PDBTitle: crystal structure of a cc-hex mutant that forms a parallel six-helix2 coiled coil cc-hex*-l24e