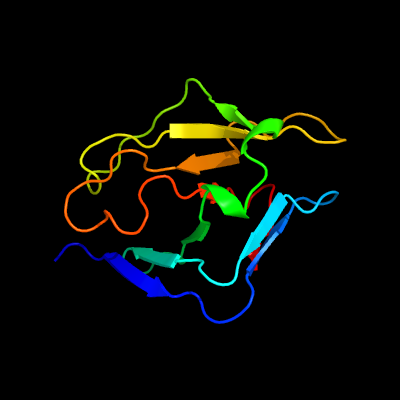
1 c3djmA_
100.0
25
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein duf427;PDBTitle: crystal structure of a protein of unknown function from duf427 family2 (rsph17029_0682) from rhodobacter sphaeroides 2.4.1 at 2.51 a3 resolution
2 d1w66a1
29.9
13
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
3 c5hj0C_
22.1
18
PDB header: ligaseChain: C: PDB Molecule: kinetochore protein mis18;PDBTitle: crystal structure of mis18 'yippee-like' domain
4 c4b1qP_
21.0
41
PDB header: toxinChain: P: PDB Molecule: conotoxin cctx;PDBTitle: nmr structure of the glycosylated conotoxin cctx from conus consors
5 c4l0zA_
20.4
42
PDB header: transcription/dnaChain: A: PDB Molecule: runt-related transcription factor 1;PDBTitle: crystal structure of runx1 and ets1 bound to tcr alpha promoter2 (crystal form 2)
6 c2mjcA_
14.1
67
PDB header: metal binding proteinChain: A: PDB Molecule: eukaryotic translation initiation factor 3 subunit g;PDBTitle: zn-binding domain of eukaryotic translation initiation factor 3,2 subunit g
7 c3icoA_
12.8
20
PDB header: hydrolaseChain: A: PDB Molecule: 6-phosphogluconolactonase;PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 tuberculosis
8 d1w9sa_
12.0
31
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM6
9 c5o60X_
11.7
33
PDB header: ribosomeChain: X: PDB Molecule: 50s ribosomal protein l27;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
10 c1z8rA_
11.7
44
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
11 c4lmgD_
10.1
33
PDB header: transcription activator/dnaChain: D: PDB Molecule: iron-regulated transcriptional activator aft2;PDBTitle: crystal structure of aft2 in complex with dna
12 c4azzB_
10.1
11
PDB header: hydrolaseChain: B: PDB Molecule: levanase;PDBTitle: carbohydrate binding module cbm66 from bacillus subtilis
13 c4e6nB_
9.7
19
PDB header: protein bindingChain: B: PDB Molecule: methyltransferase type 12;PDBTitle: crystal structure of bacterial pnkp-c/hen1-n heterodimer
14 c6oswA_
9.3
13
PDB header: cell cycleChain: A: PDB Molecule: forkhead box m1;PDBTitle: an order-to-disorder structural switch activates the foxm12 transcription factor
15 c4cawA_
9.1
26
PDB header: transferaseChain: A: PDB Molecule: glycylpeptide n-tetradecanoyltransferase;PDBTitle: crystal structure of aspergillus fumigatus n-myristoyl2 transferase in complex with myristoyl coa and a pyrazole3 sulphonamide ligand
16 c5c17A_
8.9
24
PDB header: lyaseChain: A: PDB Molecule: merb2;PDBTitle: crystal structure of the mercury-bound form of merb2
17 c2kd2A_
8.9
16
PDB header: apoptosisChain: A: PDB Molecule: fas apoptotic inhibitory molecule 1;PDBTitle: nmr structure of faim-ctd
18 d1txna_
7.9
40
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase
19 d1ne7a_
7.8
16
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: NagB-like
20 d1m7ja2
7.8
26
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: D-aminoacylase
21 c3oc6A_
not modelled
7.7
22
PDB header: hydrolaseChain: A: PDB Molecule: 6-phosphogluconolactonase;PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 smegmatis, apo form
22 d1iica2
not modelled
7.3
22
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT
23 c6ch0I_
not modelled
7.2
15
PDB header: hydrolaseChain: I: PDB Molecule: beta-lactamase;PDBTitle: structure of the quorum quenching lactonase from alicyclobacillus2 acidoterrestris bound to a glycerol molecule
24 d1vr3a1
not modelled
7.1
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Acireductone dioxygenase
25 d1rxta2
not modelled
6.8
13
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT
26 c2pcjB_
not modelled
6.8
12
PDB header: hydrolaseChain: B: PDB Molecule: lipoprotein-releasing system atp-binding protein lold;PDBTitle: crystal structure of abc transporter (aq_297) from aquifex aeolicus2 vf5
27 d1wh2a_
not modelled
6.7
18
Fold: GYF/BRK domain-likeSuperfamily: GYF domainFamily: GYF domain
28 d2cu2a1
not modelled
6.7
13
Fold: Single-stranded left-handed beta-helixSuperfamily: Guanosine diphospho-D-mannose pyrophosphorylase/mannose-6-phosphate isomerase linker domainFamily: Guanosine diphospho-D-mannose pyrophosphorylase/mannose-6-phosphate isomerase linker domain
29 d2a1fa1
not modelled
6.6
17
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like
30 d1z9da1
not modelled
6.5
22
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like
31 d1iyka2
not modelled
6.2
12
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT
32 c2kl4A_
not modelled
6.2
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2032 protein;PDBTitle: nmr structure of the protein nb7804a
33 c2j0eA_
not modelled
6.1
20
PDB header: hydrolaseChain: A: PDB Molecule: 6-phosphogluconolactonase;PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
34 d1tkla_
not modelled
6.1
40
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase
35 c5eo6A_
not modelled
6.1
40
PDB header: oxidoreductaseChain: A: PDB Molecule: coproporphyrinogen oxidase;PDBTitle: coproporphyrinogen iii oxidase (hemf) from acinetobacter baumannii
36 d1boba_
not modelled
6.0
27
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
37 c3zf7t_
not modelled
5.9
27
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
38 d2j44a2
not modelled
5.9
9
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: PUD-like
39 c3cjsA_
not modelled
5.8
37
PDB header: transferase/ribosomal proteinChain: A: PDB Molecule: ribosomal protein l11 methyltransferase;PDBTitle: minimal recognition complex between prma and ribosomal protein l11
40 c3m9bK_
not modelled
5.7
21
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
41 c2aexA_
not modelled
5.6
40
PDB header: oxidoreductaseChain: A: PDB Molecule: coproporphyrinogen iii oxidase, mitochondrial;PDBTitle: the 1.58a crystal structure of human coproporphyrinogen oxidase2 reveals the structural basis of hereditary coproporphyria
42 c5xyiL_
not modelled
5.6
44
PDB header: ribosomeChain: L: PDB Molecule: uncharacterized protein;PDBTitle: small subunit of trichomonas vaginalis ribosome
43 c6fhtB_
not modelled
5.5
27
PDB header: lyaseChain: B: PDB Molecule: bacteriophytochrome,adenylate cyclase;PDBTitle: crystal structure of an artificial phytochrome regulated2 adenylate/guanylate cyclase in its dark adapted pr form
44 d1tsfa_
not modelled
5.5
25
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
45 c3zr6A_
not modelled
5.5
12
PDB header: hydrolaseChain: A: PDB Molecule: galactocerebrosidase;PDBTitle: structure of galactocerebrosidase from mouse in complex with galactose
46 c3nmbA_
not modelled
5.4
10
PDB header: hydrolaseChain: A: PDB Molecule: putative sugar hydrolase;PDBTitle: crystal structure of a putative sugar hydrolase (bacova_03189) from2 bacteroides ovatus at 2.40 a resolution
47 c3mhxB_
not modelled
5.2
8
PDB header: metal transportChain: B: PDB Molecule: putative ferrous iron transport protein a;PDBTitle: crystal structure of stenotrophomonas maltophilia feoa complexed with2 zinc: a unique procaryotic sh3 domain protein possibly acting as a3 bacterial ferrous iron transport activating factor
48 c2c26A_
not modelled
5.2
27
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structural basis for the promiscuous specificity of the2 carbohydrate-binding modules from the beta-sandwich super3 family
49 d2k49a1
not modelled
5.2
11
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like
50 d1lnga_
not modelled
5.1
28
Fold: SRP19Superfamily: SRP19Family: SRP19
51 c2xhaB_
not modelled
5.1
20
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of domain 2 of thermotoga maritima n-utilization2 substance g (nusg)