| 1 |
|
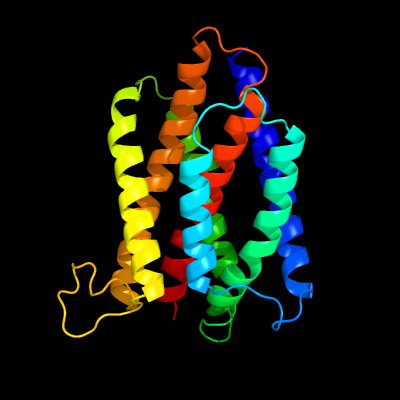
PDB 4pgu chain A
Region: 50 - 277
Aligned: 205
Modelled: 228
Confidence: 100.0%
Identity: 20%
PDB header:membrance protein
Chain: A: PDB Molecule:uncharacterized protein yetj;
PDBTitle: crystal structure of yetj from bacillus subtilis at ph 7 by soaking
Phyre2
| 2 |
|
PDB 4pgw chain A
Region: 54 - 275
Aligned: 194
Modelled: 204
Confidence: 100.0%
Identity: 19%
PDB header:membrance protein
Chain: A: PDB Molecule:uncharacterized protein yetj;
PDBTitle: crystal structure of yetj from bacillus subtilis at ph 6 by pt-sad
Phyre2
| 3 |
|
PDB 5vyl chain A
Region: 230 - 262
Aligned: 32
Modelled: 33
Confidence: 28.5%
Identity: 6%
PDB header:viral protein
Chain: A: PDB Molecule:inner tegument protein;
PDBTitle: crystal structure of n-terminal half of herpes simplex virus type 12 ul37 protein
Phyre2
| 4 |
|
PDB 4k70 chain B
Region: 230 - 262
Aligned: 32
Modelled: 33
Confidence: 20.3%
Identity: 25%
PDB header:viral protein
Chain: B: PDB Molecule:ul37;
PDBTitle: crystal structure of n-terminal half of pseudorabiesvirus ul37 protein
Phyre2
| 5 |
|
PDB 4z6y chain D
Region: 266 - 278
Aligned: 13
Modelled: 13
Confidence: 10.7%
Identity: 38%
PDB header:hydrolase inhibitor/protein binding
Chain: D: PDB Molecule:hamartin;
PDBTitle: structure of the tbc1d7-tsc1 complex
Phyre2
| 6 |
|
PDB 4z6y chain C
Region: 266 - 278
Aligned: 13
Modelled: 13
Confidence: 10.7%
Identity: 38%
PDB header:hydrolase inhibitor/protein binding
Chain: C: PDB Molecule:hamartin;
PDBTitle: structure of the tbc1d7-tsc1 complex
Phyre2
| 7 |
|
PDB 5oqt chain C
Region: 214 - 231
Aligned: 18
Modelled: 18
Confidence: 10.4%
Identity: 6%
PDB header:transport protein
Chain: C: PDB Molecule:uncharacterized protein ynem;
PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue
Phyre2
| 8 |
|
PDB 4z6y chain F
Region: 266 - 278
Aligned: 13
Modelled: 13
Confidence: 9.9%
Identity: 38%
PDB header:hydrolase inhibitor/protein binding
Chain: F: PDB Molecule:hamartin;
PDBTitle: structure of the tbc1d7-tsc1 complex
Phyre2
| 9 |
|
PDB 6f34 chain C
Region: 212 - 231
Aligned: 20
Modelled: 20
Confidence: 9.6%
Identity: 10%
PDB header:membrane protein
Chain: C: PDB Molecule:mgts;
PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue bound to arginine.
Phyre2
| 10 |
|
PDB 6hwh chain W
Region: 105 - 269
Aligned: 164
Modelled: 165
Confidence: 9.4%
Identity: 12%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase subunit 3;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
Phyre2
| 11 |
|
PDB 1m47 chain A
Region: 229 - 259
Aligned: 31
Modelled: 31
Confidence: 9.2%
Identity: 13%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Short-chain cytokines
Phyre2
| 12 |
|
PDB 5ejc chain C
Region: 266 - 275
Aligned: 10
Modelled: 10
Confidence: 8.7%
Identity: 50%
PDB header:signaling protein
Chain: C: PDB Molecule:hamartin;
PDBTitle: crystal structural of the tsc1-tbc1d7 complex
Phyre2
| 13 |
|
PDB 6ith chain A
Region: 141 - 166
Aligned: 26
Modelled: 26
Confidence: 7.8%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:syndecan-2;
PDBTitle: structure of the transmembrane domain of syndecan 2 in micelles
Phyre2
| 14 |
|
PDB 5z1l chain L
Region: 141 - 161
Aligned: 21
Modelled: 21
Confidence: 6.4%
Identity: 14%
PDB header:protein fibril
Chain: L: PDB Molecule:flagellin;
PDBTitle: cryo-em structure of methanoccus maripaludis archaellum
Phyre2
| 15 |
|
PDB 1v54 chain C
Region: 113 - 269
Aligned: 155
Modelled: 157
Confidence: 5.6%
Identity: 12%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2