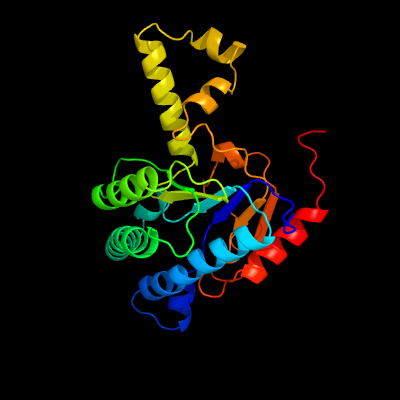
| 1 | c2vg2C_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: C: PDB Molecule:undecaprenyl pyrophosphate synthetase;
PDBTitle: rv2361 with ipp
|
|
|
|
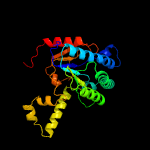
| 2 | c4h8eA_
|
|
 |
100.0 |
30 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of s. aureus undecaprenyl diphosphate synthase in complex2 with fpp and sulfate
|
|
|
|
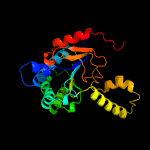
| 3 | c2vfwB_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:short-chain z-isoprenyl diphosphate synthetase;
PDBTitle: rv1086 native
|
|
|
|
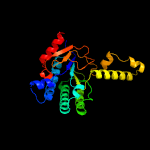
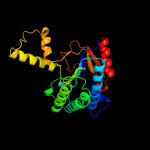
| 4 | c5hc7A_
|
|
 |
100.0 |
32 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:prenyltransference for protein;
PDBTitle: crystal structure of lavandulyl diphosphate synthase from lavandula x2 intermedia in complex with s-thiolo-isopentenyldiphosphate
|
|
|
|
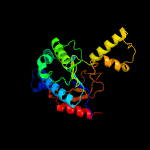
| 5 | c4q9mA_
|
|
 |
100.0 |
33 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:isoprenyl transferase;
PDBTitle: crystal structure of upps in complex with fpp and an allosteric2 inhibitor
|
|
|
|
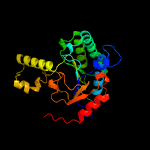
| 6 | c5hxpA_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:(2z,6z)-farnesyl diphosphate synthase, chloroplastic;
PDBTitle: crystal structure of z,z-farnesyl diphosphate synthase (d71m, e75a and2 h103y mutants) complexed with ipp
|
|
|
|
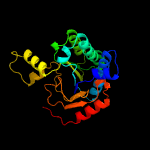
| 7 | c5gukA_
|
|
 |
100.0 |
28 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cyclolavandulyl diphosphate synthase;
PDBTitle: crystal structure of apo form of cyclolavandulyl diphosphate synthase2 (clds) from streptomyces sp. cl190
|
|
|
|
| 8 | d1ueha_
|
|
 |
100.0 |
33 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
|
| 9 | c6acsA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:ditrans,polycis-undecaprenyl-diphosphate synthase ((2e,6e)-
PDBTitle: poly-cis-prenyltransferase
|
|
|
|
| 10 | c5xk9F_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: F: PDB Molecule:undecaprenyl diphosphate synthase;
PDBTitle: crystal structure of isosesquilavandulyl diphosphate synthase from2 streptomyces sp. strain cnh-189 in complex with gspp and dmapp
|
|
|
|
| 11 | c1jp3A_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of e.coli undecaprenyl pyrophosphate synthase
|
|
|
|
| 12 | d1f75a_
|
|
 |
100.0 |
30 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
|
| 13 | c3ugsB_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of a probable undecaprenyl diphosphate synthase2 (upps) from campylobacter jejuni
|
|
|
|
| 14 | c2d2rA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
|
|
|
|
| 15 | c6jcnB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:dehydrodolichyl diphosphate synthase complex subunit nus1;
PDBTitle: yeast dehydrodolichyl diphosphate synthase complex subunit nus1
|
|
|
|
| 16 | c4cmxB_
|
|
 |
99.3 |
18 |
PDB header:nuclear protein
Chain: B: PDB Molecule:rv3378c;
PDBTitle: crystal structure of rv3378c
|
|
|
|
| 17 | c3cprB_
|
|
 |
70.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
|
|
|
| 18 | c3s5oA_
|
|
 |
64.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
|
|
|
| 19 | c5ak1A_
|
|
 |
62.7 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate binding family 6;
PDBTitle: the penta-modular cellulosomal arabinoxylanase ctxyl5a2 structure as revealed by x-ray crystallography
|
|
|
|
| 20 | c3fluD_
|
|
 |
58.2 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
|
|
|
| 21 | c3n58D_ |
|
not modelled |
58.1 |
8 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
|
|
| 22 | c2rfgB_ |
|
not modelled |
58.1 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
|
|
| 23 | c3si9B_ |
|
not modelled |
57.1 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
|
|
| 24 | c5n6yD_ |
|
not modelled |
56.8 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase vanadium-iron protein alpha chain;
PDBTitle: azotobacter vinelandii vanadium nitrogenase
|
|
|
| 25 | c2ou4C_ |
|
not modelled |
56.6 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
|
|
| 26 | c3daqB_ |
|
not modelled |
55.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
|
|
| 27 | c5iq5A_ |
|
not modelled |
52.8 |
17 |
PDB header:viral protein
Chain: A: PDB Molecule:macro domain;
PDBTitle: nmr solution structure of mayaro virus macro domain
|
|
|
| 28 | d1hl2a_ |
|
not modelled |
51.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 29 | c2hk1D_ |
|
not modelled |
50.3 |
14 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
|
|
| 30 | c3pueA_ |
|
not modelled |
49.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
| 31 | c3zddA_ |
|
not modelled |
49.7 |
14 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:protein xni;
PDBTitle: structure of e. coli exoix in complex with the palindromic 5ov62 oligonucleotide and potassium
|
|
|
| 32 | d2a6na1 |
|
not modelled |
49.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 33 | c6c34A_ |
|
not modelled |
48.8 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:5'-3' exonuclease;
PDBTitle: mycobacterium smegmatis dna flap endonuclease mutant d125n
|
|
|
| 34 | c5ud6B_ |
|
not modelled |
47.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
| 35 | c2v9dB_ |
|
not modelled |
47.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
|
|
| 36 | c3qfeB_ |
|
not modelled |
47.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
|
|
| 37 | d1xxxa1 |
|
not modelled |
46.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 38 | c3bdkB_ |
|
not modelled |
46.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
|
|
| 39 | c3noeA_ |
|
not modelled |
46.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
|
|
| 40 | c2yxgD_ |
|
not modelled |
45.8 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
|
|
| 41 | c3fkkA_ |
|
not modelled |
45.2 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 42 | c3dx5A_ |
|
not modelled |
45.0 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
|
|
| 43 | d1i60a_ |
|
not modelled |
44.0 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 44 | c3d0cB_ |
|
not modelled |
43.6 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 45 | c2vc6A_ |
|
not modelled |
43.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
|
|
| 46 | c3n2xB_ |
|
not modelled |
41.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
|
|
| 47 | c3b4uB_ |
|
not modelled |
39.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
|
|
| 48 | c3lciA_ |
|
not modelled |
38.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
|
|
| 49 | c3g0sA_ |
|
not modelled |
38.3 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
|
|
| 50 | c3cnyA_ |
|
not modelled |
37.8 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
|
|
| 51 | c3cqkB_ |
|
not modelled |
36.5 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
|
|
| 52 | d1o5ka_ |
|
not modelled |
35.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 53 | d1xkya1 |
|
not modelled |
35.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 54 | c4i7vD_ |
|
not modelled |
35.0 |
15 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: agrobacterium tumefaciens dhdps with pyruvate
|
|
|
| 55 | c5afdA_ |
|
not modelled |
34.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: native structure of n-acetylneuramininate lyase (sialic acid aldolase)2 from aliivibrio salmonicida
|
|
|
| 56 | c4ah7C_ |
|
not modelled |
34.6 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
|
|
|
| 57 | c3qxbB_ |
|
not modelled |
34.1 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
|
|
| 58 | c3lerA_ |
|
not modelled |
32.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
|
|
|
| 59 | c5ktlA_ |
|
not modelled |
31.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 60 | c5ui3C_ |
|
not modelled |
31.7 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from chlamydomonas reinhardtii
|
|
|
| 61 | c3gqeA_ |
|
not modelled |
30.0 |
15 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of venezuelan equine encephalitis2 virus
|
|
|
| 62 | c4iqyB_ |
|
not modelled |
28.9 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:o-acetyl-adp-ribose deacetylase macrod2;
PDBTitle: crystal structure of the human protein-proximal adp-ribosyl-hydrolase2 macrod2
|
|
|
| 63 | c3oneA_ |
|
not modelled |
28.7 |
11 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
|
|
| 64 | c3eb2A_ |
|
not modelled |
28.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
|
|
| 65 | c3bi8A_ |
|
not modelled |
28.4 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
|
|
| 66 | c5c54D_ |
|
not modelled |
27.9 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase/n-acetylneuraminate lyase;
PDBTitle: crystal structure of a novel n-acetylneuraminic acid lyase from2 corynebacterium glutamicum
|
|
|
| 67 | c4nq1B_ |
|
not modelled |
27.8 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 68 | c6mqhA_ |
|
not modelled |
27.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate synthase (htpa2 synthase) from burkholderia mallei
|
|
|
| 69 | c6d0gA_ |
|
not modelled |
27.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pirin family protein;
PDBTitle: 1.78 angstrom resolution crystal structure of quercetin 2,3-2 dioxygenase from acinetobacter baumannii
|
|
|
| 70 | c4umlA_ |
|
not modelled |
25.5 |
4 |
PDB header:signaling protein
Chain: A: PDB Molecule:ganglioside-induced differentiation-associated protein 2;
PDBTitle: crystal structure of ganglioside induced differentiation2 associated protein 2 (gdap2) macro domain
|
|
|
| 71 | c2x47A_ |
|
not modelled |
25.5 |
7 |
PDB header:signaling protein
Chain: A: PDB Molecule:macro domain-containing protein 1;
PDBTitle: crystal structure of human macrod1
|
|
|
| 72 | c6daqA_ |
|
not modelled |
25.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:phdj;
PDBTitle: phdj bound to substrate intermediate
|
|
|
| 73 | c2ynmD_ |
|
not modelled |
25.0 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
|
|
|
| 74 | d1vk1a_ |
|
not modelled |
24.9 |
38 |
Fold:ParB/Sulfiredoxin
Superfamily:ParB/Sulfiredoxin
Family:Hypothetical protein PF0380 |
|
|
| 75 | d1miob_ |
|
not modelled |
24.1 |
9 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 76 | c4ur7B_ |
|
not modelled |
23.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:keto-deoxy-d-galactarate dehydratase;
PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
|
|
|
| 77 | c2f9iC_ |
|
not modelled |
23.2 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: crystal structure of the carboxyltransferase subunit of acc from2 staphylococcus aureus
|
|
|
| 78 | c2ksnA_ |
|
not modelled |
22.8 |
36 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin domain-containing protein 2;
PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
|
|
|
| 79 | d1cmwa2 |
|
not modelled |
22.8 |
15 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 80 | c1cmwA_ |
|
not modelled |
22.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:protein (dna polymerase i);
PDBTitle: crystal structure of taq dna-polymerase shows a new orientation for2 the structure-specific nuclease domain
|
|
|
| 81 | d2f9ya1 |
|
not modelled |
21.9 |
12 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
|
|
| 82 | c4uxdC_ |
|
not modelled |
21.9 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|
| 83 | c4xkyC_ |
|
not modelled |
21.5 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
|
|
|
| 84 | c3d64A_ |
|
not modelled |
21.4 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
|
|
| 85 | c4icnB_ |
|
not modelled |
21.2 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from shewanella benthica
|
|
|
| 86 | c2d1gB_ |
|
not modelled |
21.1 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:acid phosphatase;
PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
|
|
|
| 87 | c2r8wB_ |
|
not modelled |
20.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
|
|
| 88 | c3vylB_ |
|
not modelled |
20.2 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose 3-epimerase;
PDBTitle: structure of l-ribulose 3-epimerase
|
|
|
| 89 | c5n6yE_ |
|
not modelled |
19.9 |
11 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nitrogenase vanadium-iron protein beta chain;
PDBTitle: azotobacter vinelandii vanadium nitrogenase
|
|
|
| 90 | c2pfuA_ |
|
not modelled |
19.8 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:biopolymer transport exbd protein;
PDBTitle: nmr strcuture determination of the periplasmic domain of exbd from2 e.coli
|
|
|
| 91 | c5by4A_ |
|
not modelled |
19.6 |
24 |
PDB header:protein transport
Chain: A: PDB Molecule:protein tolr;
PDBTitle: structure and function of the escherichia coli tol-pal stator protein2 tolr
|
|
|
| 92 | c2nuxB_ |
|
not modelled |
18.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
|
|
| 93 | c5v96A_ |
|
not modelled |
18.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:s-adenosyl-l-homocysteine hydrolase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 naegleria fowleri with bound nad and adenosine
|
|
|
| 94 | c3ktcB_ |
|
not modelled |
18.6 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
|
|
| 95 | c5j67C_ |
|
not modelled |
18.5 |
20 |
PDB header:membrane protein
Chain: C: PDB Molecule:astrotactin-2;
PDBTitle: structure of astrotactin-2, a conserved vertebrate-specific and2 perforin-like membrane protein involved in neuronal development
|
|
|
| 96 | c5ujeA_ |
|
not modelled |
18.5 |
23 |
PDB header:gene regulation
Chain: A: PDB Molecule:sbni protein;
PDBTitle: sbni with c-terminal truncation from staphylococcus aureus
|
|
|
| 97 | c5kivA_ |
|
not modelled |
18.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-adp-ribose hydrolase;
PDBTitle: crystal structure of saumacro (sav0325)
|
|
|
| 98 | c3vniC_ |
|
not modelled |
18.0 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 99 | c5hm8C_ |
|
not modelled |
17.5 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: 2.85 angstrom crystal structure of s-adenosylhomocysteinase from2 cryptosporidium parvum in complex with adenosine and nad.
|
|
|