| 1 |
|
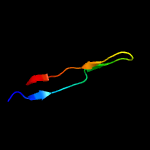
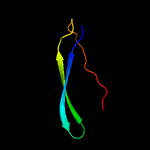
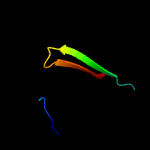
PDB 1heh chain C
Region: 61 - 84
Aligned: 24
Modelled: 24
Confidence: 94.2%
Identity: 13%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family II
Phyre2
| 2 |
|
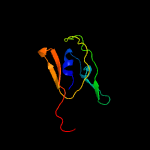
PDB 1e5b chain A
Region: 62 - 84
Aligned: 23
Modelled: 23
Confidence: 94.0%
Identity: 26%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family II
Phyre2
| 3 |
|
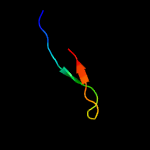
PDB 6f7e chain A
Region: 61 - 101
Aligned: 41
Modelled: 41
Confidence: 93.5%
Identity: 24%
PDB header:carbohydrate
Chain: A: PDB Molecule:putative secreted cellulose binding protein;
PDBTitle: nmr solution structure of the cellulose-binding family 2 carbohydrate2 binding domain (cbm2) from sclpmo9c
Phyre2
| 4 |
|
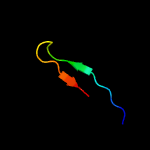
PDB 3ndy chain G
Region: 62 - 101
Aligned: 40
Modelled: 40
Confidence: 91.0%
Identity: 23%
PDB header:hydrolase
Chain: G: PDB Molecule:endoglucanase d;
PDBTitle: the structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
Phyre2
| 5 |
|
PDB 2rtt chain A
Region: 62 - 101
Aligned: 40
Modelled: 40
Confidence: 89.7%
Identity: 25%
PDB header:hydrolase
Chain: A: PDB Molecule:chic;
PDBTitle: solution structure of the chitin-binding domain of chi18ac from2 streptomyces coelicolor
Phyre2
| 6 |
|
PDB 1exh chain A
Region: 56 - 100
Aligned: 44
Modelled: 45
Confidence: 89.7%
Identity: 23%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family II
Phyre2
| 7 |
|
PDB 2cwr chain A
Region: 64 - 84
Aligned: 20
Modelled: 21
Confidence: 73.4%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase;
PDBTitle: crystal structure of chitin biding domain of chitinase from2 pyrococcus furiosus
Phyre2
| 8 |
|
PDB 3icg chain D
Region: 55 - 84
Aligned: 30
Modelled: 30
Confidence: 56.0%
Identity: 30%
PDB header:hydrolase
Chain: D: PDB Molecule:endoglucanase d;
PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
Phyre2
| 9 |
|
PDB 3hbz chain A
Region: 17 - 93
Aligned: 77
Modelled: 77
Confidence: 31.1%
Identity: 23%
PDB header:ca-binding protein
Chain: A: PDB Molecule:putative glycoside hydrolase;
PDBTitle: crystal structure of a putative glycoside hydrolase (bt_2081) from2 bacteroides thetaiotaomicron vpi-5482 at 2.05 a resolution
Phyre2
| 10 |
|
PDB 1h2d chain A
Region: 79 - 96
Aligned: 18
Modelled: 18
Confidence: 23.0%
Identity: 44%
PDB header:virus/viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: ebola virus matrix protein vp40 n-terminal domain in complex with rna2 (low-resolution vp40[31-212] variant).
Phyre2
| 11 |
|
PDB 1na6 chain A domain 1
Region: 75 - 91
Aligned: 17
Modelled: 17
Confidence: 22.9%
Identity: 29%
Fold: DNA-binding pseudobarrel domain
Superfamily: DNA-binding pseudobarrel domain
Family: Type II restriction endonuclease effector domain
Phyre2
| 12 |
|
PDB 1h2c chain A
Region: 79 - 96
Aligned: 18
Modelled: 18
Confidence: 22.9%
Identity: 44%
Fold: EV matrix protein
Superfamily: EV matrix protein
Family: EV matrix protein
Phyre2
| 13 |
|
PDB 3tcq chain A
Region: 79 - 88
Aligned: 10
Modelled: 10
Confidence: 20.9%
Identity: 70%
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: crystal structure of matrix protein vp40 from ebola virus sudan
Phyre2
| 14 |
|
PDB 4het chain A
Region: 17 - 93
Aligned: 77
Modelled: 77
Confidence: 18.7%
Identity: 22%
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical glycoside hydrolase;
PDBTitle: crystal structure of a putative glycoside hydrolase (bt3745) from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
Phyre2
| 15 |
|
PDB 1es6 chain A domain 1
Region: 79 - 96
Aligned: 18
Modelled: 18
Confidence: 16.8%
Identity: 44%
Fold: EV matrix protein
Superfamily: EV matrix protein
Family: EV matrix protein
Phyre2
| 16 |
|
PDB 1es6 chain A
Region: 79 - 96
Aligned: 18
Modelled: 18
Confidence: 13.3%
Identity: 44%
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: crystal structure of the matrix protein of ebola virus
Phyre2
| 17 |
|
PDB 6ekv chain A
Region: 29 - 55
Aligned: 27
Modelled: 27
Confidence: 12.6%
Identity: 41%
PDB header:lipid binding protein
Chain: A: PDB Molecule:toxin complex component orf-x2;
PDBTitle: structure of orfx2 from clostridium botulinum a2
Phyre2
| 18 |
|
PDB 5jk2 chain I
Region: 70 - 77
Aligned: 8
Modelled: 8
Confidence: 10.6%
Identity: 63%
PDB header:cell adhesion
Chain: I: PDB Molecule:tp0751;
PDBTitle: crystal structure of treponema pallidum tp0751 (pallilysin)
Phyre2
| 19 |
|
PDB 5b0v chain A
Region: 79 - 88
Aligned: 10
Modelled: 10
Confidence: 10.1%
Identity: 60%
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: crystal structure of marburg virus vp40 dimer
Phyre2
| 20 |
|
PDB 3ogi chain C
Region: 83 - 90
Aligned: 8
Modelled: 8
Confidence: 6.9%
Identity: 75%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative esat-6-like protein 6;
PDBTitle: crystal structure of the mycobacterium tuberculosis h37rv esxop2 complex (rv2346c-rv2347c)
Phyre2