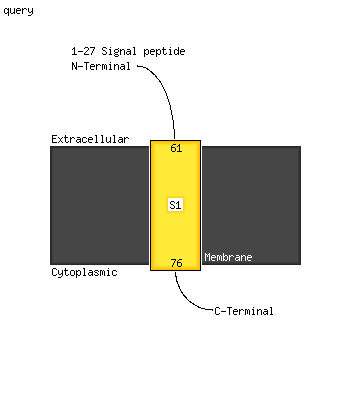
1 c2c1iA_
100.0
36
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan glcnac deacetylase;PDBTitle: structure of streptococcus pneumoniae peptidoglycan deacetylase2 (sppgda) d 275 n mutant.
2 c5jp6A_
100.0
29
PDB header: hydrolaseChain: A: PDB Molecule: putative polysaccharide deacetylase;PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
3 c4nz3A_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: deacetylase da1;PDBTitle: structure of vibrio cholerae chitin de-n-acetylase in complex with2 di(n-acetyl-d-glucosamine) (cbs) in p 21 21 21
4 c5lgcA_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: arce4a;PDBTitle: t48 deacetylase with substrate
5 c1w17A_
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: probable polysaccharide deacetylase pdaa;PDBTitle: structure of bacillus subtilis pdaa, a family 4 carbohydrate esterase.
6 d1ny1a_
100.0
28
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
7 d2cc0a1
100.0
36
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
8 d2c1ia1
100.0
37
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
9 c5ncdA_
100.0
31
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan n-acetylglucosamine deacetylase;PDBTitle: crystal structure of the polysaccharide deacetylase bc1974 from2 bacillus cereus in complex with (2s)-2-amino-5-3 (diaminomethylideneamino)-n-hydroxypentanamide
10 c4m1bA_
100.0
26
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: structural determination of ba0150, a polysaccharide deacetylase from2 bacillus anthracis
11 d2j13a1
100.0
27
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
12 c2vyoA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase domain-containing proteinPDBTitle: chitin deacetylase family member from encephalitozoon cuniculi
13 c2y8uA_
100.0
31
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: a. nidulans chitin deacetylase
14 c4l1gB_
100.0
30
PDB header: hydrolaseChain: B: PDB Molecule: peptidoglycan n-acetylglucosamine deacetylase;PDBTitle: crystal structure of the bc1960 peptidoglycan n-acetylglucosamine2 deacetylase from bacillus cereus
15 d2iw0a1
100.0
32
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
16 d2c71a1
100.0
31
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
17 c2iw0A_
100.0
32
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: structure of the chitin deacetylase from the fungal2 pathogen colletotrichum lindemuthianum
18 c2w3zA_
100.0
26
PDB header: hydrolaseChain: A: PDB Molecule: putative deacetylase;PDBTitle: structure of a streptococcus mutans ce4 esterase
19 d2nlya1
100.0
19
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: Divergent polysaccharide deacetylase
20 c3qbuD_
100.0
25
PDB header: hydrolaseChain: D: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of putative peptidoglycan deactelyase (hp0310) from2 helicobacter pylori
21 d1z7aa1
not modelled
100.0
16
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: PA1517-like
22 c3rxzA_
not modelled
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: crystal structure of putative polysaccharide deacetylase from2 mycobacterium smegmatis
23 c3s6oD_
not modelled
100.0
18
PDB header: hydrolaseChain: D: PDB Molecule: polysaccharide deacetylase family protein;PDBTitle: crystal structure of a polysaccharide deacetylase family protein from2 burkholderia pseudomallei
24 c5z34A_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: the structure of a chitin deacetylase from bombyx mori provide the2 first insight into insect chitin deacetylation mechanism
25 c6dq3B_
not modelled
100.0
25
PDB header: hydrolaseChain: B: PDB Molecule: polysaccharide deacetylase;PDBTitle: streptococcus pyogenes deacetylase pdi in complex with acetate
26 c5znsA_
not modelled
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: insect chitin deacetylase
27 c4wcjA_
not modelled
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: structure of icab from ammonifex degensii
28 c4f9dA_
not modelled
99.9
20
PDB header: hydrolaseChain: A: PDB Molecule: poly-beta-1,6-n-acetyl-d-glucosamine n-deacetylase;PDBTitle: structure of escherichia coli pgab 42-655 in complex with nickel
29 c5bu6B_
not modelled
99.9
23
PDB header: hydrolaseChain: B: PDB Molecule: bpsb (pgab), poly-beta-1,6-n-acetyl-d-glucosamine n-PDBTitle: structure of bpsb deaceylase domain from bordetella bronchiseptica
30 c4u10B_
not modelled
99.9
28
PDB header: hydrolaseChain: B: PDB Molecule: poly-beta-1,6-n-acetyl-d-glucosamine n-deacetylase;PDBTitle: probing the structure and mechanism of de-n-acetylase from2 aggregatibacter actinomycetemcomitans
31 c6go1A_
not modelled
99.8
28
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase-like protein;PDBTitle: crystal structure of a bacillus anthracis peptidoglycan deacetylase
32 c4hd5A_
not modelled
99.8
24
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: crystal structure of bc0361, a polysaccharide deacetylase from2 bacillus cereus
33 d1k1xa3
not modelled
98.8
19
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: 4-alpha-glucanotransferase, N-terminal domain
34 c2qv5A_
not modelled
98.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2773;PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
35 d2b5dx2
not modelled
98.6
18
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: AmyC N-terminal domain-like
36 c2b5dX_
not modelled
98.5
18
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of the novel alpha-amylase amyc from thermotoga2 maritima
37 c1k1yA_
not modelled
98.4
18
PDB header: transferaseChain: A: PDB Molecule: 4-alpha-glucanotransferase;PDBTitle: crystal structure of thermococcus litoralis 4-alpha-glucanotransferase2 complexed with acarbose
38 c5wu7A_
not modelled
98.2
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of gh57-type branching enzyme from hyperthermophilic2 archaeon pyrococcus horikoshii
39 d1ufaa2
not modelled
98.0
22
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: AmyC N-terminal domain-like
40 c1ufaA_
not modelled
98.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tt1467 protein;PDBTitle: crystal structure of tt1467 from thermus thermophilus hb8
41 c3n92A_
not modelled
97.8
19
PDB header: transferaseChain: A: PDB Molecule: alpha-amylase, gh57 family;PDBTitle: crystal structure of tk1436, a gh57 branching enzyme from2 hyperthermophilic archaeon thermococcus kodakaraensis, in complex3 with glucose
42 c3hftA_
not modelled
97.4
15
PDB header: hydrolaseChain: A: PDB Molecule: wbms, polysaccharide deacetylase involved in o-antigenPDBTitle: crystal structure of a putative polysaccharide deacetylase involved in2 o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica3 at 1.90 a resolution
43 d1v6ta_
not modelled
97.3
19
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like
44 d2i5ia1
not modelled
95.0
16
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: YdjC-like
45 c2wyhA_
not modelled
94.4
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase;PDBTitle: structure of the streptococcus pyogenes family gh38 alpha-2 mannosidase
46 c2ow7A_
not modelled
94.0
19
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase 2;PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
47 c1htyA_
not modelled
93.6
19
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase ii;PDBTitle: golgi alpha-mannosidase ii
48 c1o7dA_
not modelled
91.6
15
PDB header: hydrolaseChain: A: PDB Molecule: lysosomal alpha-mannosidase;PDBTitle: the structure of the bovine lysosomal a-mannosidase suggests a novel2 mechanism for low ph activation
49 d3bvua3
not modelled
91.6
17
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: alpha-mannosidase
50 d2dfaa1
not modelled
90.6
20
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like
51 c3lvtA_
not modelled
87.8
12
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase, family 38;PDBTitle: the crystal structure of a protein in the glycosyl hydrolase family 382 from enterococcus faecalis to 2.55a
52 c6b9pA_
not modelled
86.8
14
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase from canavalia ensiformis (jack bean);PDBTitle: structure of gh 38 jack bean alpha-mannosidase in complex with a 36-2 valent iminosugar cluster inhibitor
53 c4ovxA_
not modelled
85.4
11
PDB header: isomeraseChain: A: PDB Molecule: xylose isomerase domain protein tim barrel;PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
54 c5exkG_
not modelled
84.4
13
PDB header: transferaseChain: G: PDB Molecule: lipoyl synthase;PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
55 c2x5eA_
not modelled
76.2
13
PDB header: unknown functionChain: A: PDB Molecule: upf0271 protein pa4511;PDBTitle: crystal structure of the hypothetical protein pa4511 from2 pseudomonas aeruginosa
56 c5hk8A_
not modelled
75.0
24
PDB header: hydrolaseChain: A: PDB Molecule: probable pheophorbidase;PDBTitle: crystal strucutre of a methylesterase protein mes16 from arabidopsis
57 d2i7xa1
not modelled
72.7
10
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: beta-CASP RNA-metabolising hydrolases
58 c2i7xA_
not modelled
72.7
10
PDB header: rna binding protein, protein bindingChain: A: PDB Molecule: protein cft2;PDBTitle: structure of yeast cpsf-100 (ydh1p)
59 c3stxB_
not modelled
72.7
19
PDB header: hydrolaseChain: B: PDB Molecule: methylketone synthase 1;PDBTitle: crystal structure of tomato methylketone synthase i h243a variant2 complexed with beta-ketoheptanoate
60 c5dqpA_
not modelled
71.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: edta monooxygenase;PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
61 c3zoqA_
not modelled
71.1
27
PDB header: hydrolase/viral proteinChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of bsudg-p56 complex
62 d3c70a1
not modelled
68.4
25
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like
63 c3wwoA_
not modelled
68.1
25
PDB header: lyaseChain: A: PDB Molecule: (s)-hydroxynitrile lyase;PDBTitle: s-selective hydroxynitrile lyase from baliospermum montanum (apo1)
64 c3dqzB_
not modelled
68.0
25
PDB header: lyaseChain: B: PDB Molecule: alpha-hydroxynitrile lyase-like protein;PDBTitle: structure of the hydroxynitrile lyase from arabidopsis thaliana
65 d1tqha_
not modelled
67.2
20
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase/lipase
66 c5jm0A_
not modelled
64.8
11
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase,alpha-mannosidase,alpha-mannosidase;PDBTitle: structure of the s. cerevisiae alpha-mannosidase 1
67 d1e89a_
not modelled
63.6
22
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like
68 c2mhcA_
not modelled
63.5
10
PDB header: recombinationChain: A: PDB Molecule: tnpx;PDBTitle: nmr structure of the catalytic domain of the large serine resolvase2 tnpx
69 d1xkla_
not modelled
60.5
22
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hydroxynitrile lyase-like
70 c5nn7A_
not modelled
59.9
14
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: kshv uracil-dna glycosylase, apo form
71 c5esrA_
not modelled
58.9
28
PDB header: hydrolaseChain: A: PDB Molecule: haloalkane dehalogenase;PDBTitle: crystal structure of haloalkane dehalogenase (dcca) from caulobacter2 crescentus
72 c2owrD_
not modelled
58.0
19
PDB header: hydrolaseChain: D: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
73 c3gzjB_
not modelled
56.8
25
PDB header: hydrolaseChain: B: PDB Molecule: polyneuridine-aldehyde esterase;PDBTitle: crystal structure of polyneuridine aldehyde esterase complexed with2 16-epi-vellosimine
74 c3cxmA_
not modelled
56.4
19
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: leishmania naiffi uracil-dna glycosylase in complex with 5-bromouracil
75 c2gm4B_
not modelled
56.1
19
PDB header: recombination, dnaChain: B: PDB Molecule: transposon gamma-delta resolvase;PDBTitle: an activated, tetrameric gamma-delta resolvase: hin chimaera bound to2 cleaved dna
76 c3dyvA_
not modelled
55.5
22
PDB header: hydrolaseChain: A: PDB Molecule: esterase d;PDBTitle: snapshots of esterase d from lactobacillus rhamnosus:2 insights into a rotation driven catalytic mechanism
77 c6d6zA_
not modelled
54.6
12
PDB header: isomeraseChain: A: PDB Molecule: nickel-dependent lactate racemase;PDBTitle: structure of the 2nd lactate racemase homolog apoprotein from2 thermoanaerobacterium thermosaccharolyticum
78 d1jdqa_
not modelled
54.2
15
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like
79 c4y7dA_
not modelled
53.2
25
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase fold protein;PDBTitle: alpha/beta hydrolase fold protein from nakamurella multipartita
80 c4fbmA_
not modelled
53.1
16
PDB header: hydrolaseChain: A: PDB Molecule: lips lipolytic enzyme;PDBTitle: lips and lipt, two metagenome-derived lipolytic enzymes increase the2 diversity of known lipase and esterase families
81 d1xw8a_
not modelled
52.9
24
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like
82 d1fi4a2
not modelled
52.4
35
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: Mevalonate 5-diphosphate decarboxylase
83 c3tr7A_
not modelled
52.3
24
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
84 c5xksB_
not modelled
51.4
9
PDB header: hydrolaseChain: B: PDB Molecule: thermostable monoacylglycerol lipase;PDBTitle: crystal structure of monoacylglycerol lipase from thermophilic2 geobacillus sp. 12amor
85 d1m0sa1
not modelled
50.9
16
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
86 c2yjgB_
not modelled
50.3
13
PDB header: isomeraseChain: B: PDB Molecule: lactate racemase apoprotein;PDBTitle: structure of the lactate racemase apoprotein from2 thermoanaerobacterium thermosaccharolyticum
87 c2p6yA_
not modelled
50.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein vca0587;PDBTitle: x-ray structure of the protein q9km02_vibch from vibrio cholerae at2 the resolution 1.63 a. northeast structural genomics consortium3 target vcr80.
88 c3l2iB_
not modelled
49.8
14
PDB header: lyaseChain: B: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
89 d7reqa2
not modelled
49.1
15
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
90 d1hx7a_
not modelled
49.0
13
Fold: Resolvase-likeSuperfamily: Resolvase-likeFamily: gamma,delta resolvase, catalytic domain
91 c4gf5S_
not modelled
48.8
19
PDB header: transferaseChain: S: PDB Molecule: cals11;PDBTitle: crystal structure of calicheamicin methyltransferase, cals11
92 c5w4zA_
not modelled
48.4
21
PDB header: flavoproteinChain: A: PDB Molecule: riboflavin lyase;PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
93 c3b9nB_
not modelled
48.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: alkane monoxygenase;PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
94 c5uroA_
not modelled
48.2
28
PDB header: hydrolaseChain: A: PDB Molecule: predicted protein;PDBTitle: structure of a soluble epoxide hydrolase identified in trichoderma2 reesei
95 c4ycsC_
not modelled
48.0
8
PDB header: hydrolaseChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of putative lipoprotein from peptoclostridium2 difficile 630 (fragment)
96 c5h3hB_
not modelled
47.8
25
PDB header: hydrolaseChain: B: PDB Molecule: abhydrolase domain-containing protein;PDBTitle: esterase (eaest) from exiguobacterium antarcticum
97 c4rrfD_
not modelled
47.5
12
PDB header: ligaseChain: D: PDB Molecule: threonine--trna ligase;PDBTitle: editing domain of threonyl-trna synthetase from methanococcus2 jannaschii with l-ser3aa
98 d1a8qa_
not modelled
46.6
22
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase
99 c2hkeB_
not modelled
46.6
21
PDB header: lyaseChain: B: PDB Molecule: diphosphomevalonate decarboxylase, putative;PDBTitle: mevalonate diphosphate decarboxylase from trypanosoma brucei
100 c3rm3A_
not modelled
46.5
19
PDB header: hydrolaseChain: A: PDB Molecule: thermostable monoacylglycerol lipase;PDBTitle: crystal structure of monoacylglycerol lipase from bacillus sp. h257
101 c4z8zA_
not modelled
45.9
11
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the hypothetical protein from ruminiclostridium2 thermocellum atcc 27405
102 c6g80C_
not modelled
45.0
11
PDB header: transferaseChain: C: PDB Molecule: methyltransferase domain protein;PDBTitle: structure of mycobacterium hassiacum met1 from orthorhombic crystals.
103 d1va4a_
not modelled
44.8
22
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase
104 c5y51F_
not modelled
44.7
25
PDB header: hydrolaseChain: F: PDB Molecule: pyrethroid hydrolase;PDBTitle: crystal structure of pyth_h230a
105 c4k3zA_
not modelled
44.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
106 c4f0jA_
not modelled
44.4
28
PDB header: hydrolaseChain: A: PDB Molecule: probable hydrolytic enzyme;PDBTitle: crystal structure of a probable hydrolytic enzyme (pa3053) from2 pseudomonas aeruginosa pao1 at 1.50 a resolution
107 c3h5oB_
not modelled
44.0
9
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator gntr;PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
108 c4df3B_
not modelled
43.9
18
PDB header: transferaseChain: B: PDB Molecule: fibrillarin-like rrna/trna 2'-o-methyltransferase;PDBTitle: crystal structure of aeropyrum pernix fibrillarin in complex with2 natively bound s-adenosyl-l-methionine at 1.7a
109 c4zwnD_
not modelled
43.8
26
PDB header: hydrolaseChain: D: PDB Molecule: monoglyceride lipase;PDBTitle: crystal structure of a soluble variant of the monoglyceride lipase2 from saccharomyces cerevisiae
110 c4rpcA_
not modelled
43.1
22
PDB header: hydrolaseChain: A: PDB Molecule: putative alpha/beta hydrolase;PDBTitle: crystal structure of the putative alpha/beta hydrolase family protein2 from desulfitobacterium hafniense
111 d1brta_
not modelled
43.1
28
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase
112 c2cjpA_
not modelled
42.7
19
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: structure of potato (solanum tuberosum) epoxide hydrolase i (steh1)
113 c5xmdA_
not modelled
41.8
25
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase a;PDBTitle: crystal structure of epoxide hydrolase vreh1 from vigna radiata
114 c3hazA_
not modelled
41.7
9
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
115 c2r0qF_
not modelled
41.6
15
PDB header: recombination/dnaChain: F: PDB Molecule: putative transposon tn552 dna-invertase bin3;PDBTitle: crystal structure of a serine recombinase- dna regulatory2 complex
116 d1jr2a_
not modelled
41.2
10
Fold: HemD-likeSuperfamily: HemD-likeFamily: HemD-like
117 c1jr2A_
not modelled
41.2
10
PDB header: lyaseChain: A: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: structure of uroporphyrinogen iii synthase
118 d1b6ga_
not modelled
41.1
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase
119 c5oluA_
not modelled
40.8
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase family protein;PDBTitle: the crystal structure of a highly thermostable carboxyl esterase from2 bacillus coagulans in complex with glycerol
120 d1a8sa_
not modelled
40.6
25
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase