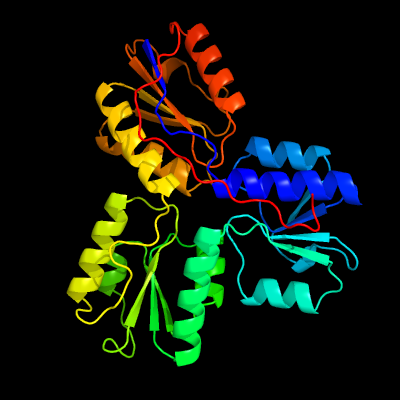
| 1 | c3urkA_
|
|
 |
100.0 |
53 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: isph in complex with propynyl diphosphate (1061)
|
|
|
|
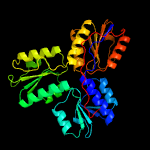
| 2 | c3ke8A_
|
|
 |
100.0 |
53 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: crystal structure of isph:hmbpp-complex
|
|
|
|
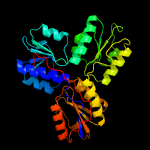
| 3 | c4n7bA_
|
|
 |
100.0 |
41 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lytb;
PDBTitle: structure of the e-1-hydroxy-2-methyl-but-2-enyl-4-diphosphate2 reductase from plasmodium falciparum
|
|
|
|
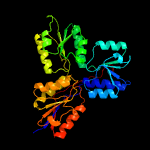
| 4 | c3dnfB_
|
|
 |
100.0 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
|
|
|
| 5 | c5c4nD_
|
|
 |
89.2 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:precorrin-6a reductase;
PDBTitle: cobk precorrin-6a reductase
|
|
|
|
| 6 | c3rfuC_
|
|
 |
87.6 |
23 |
PDB header:hydrolase, membrane protein
Chain: C: PDB Molecule:copper efflux atpase;
PDBTitle: crystal structure of a copper-transporting pib-type atpase
|
|
|
|
| 7 | c4zhtB_
|
|
 |
86.3 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:bifunctional udp-n-acetylglucosamine 2-epimerase/n-
PDBTitle: crystal structure of udp-glcnac 2-epimerase
|
|
|
|
| 8 | c3ff4A_
|
|
 |
85.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
|
|
|
| 9 | c3j09A_
|
|
 |
80.2 |
16 |
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
|
|
|
| 10 | c4zo9B_
|
|
 |
80.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:lin1840 protein;
PDBTitle: crystal structure of mutant (d270a) beta-glucosidase from listeria2 innocua in complex with laminaribiose
|
|
|
|
| 11 | c3h5lB_
|
|
 |
78.1 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc transporter;
PDBTitle: crystal structure of a putative branched-chain amino acid abc2 transporter from silicibacter pomeroyi
|
|
|
|
| 12 | c3j08A_
|
|
 |
78.0 |
16 |
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
|
|
|
| 13 | d1m2ka_
|
|
 |
77.9 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
|
|
|
| 14 | d1j6ua1
|
|
 |
76.0 |
16 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
|
| 15 | c6oviA_
|
|
 |
75.9 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
|
| 16 | c4es6A_
|
|
 |
75.2 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of hemd (pa5259) from pseudomonas aeruginosa (pao1)2 at 2.22 a resolution
|
|
|
|
| 17 | c3hutA_
|
|
 |
73.5 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from rhodospirillum rubrum
|
|
|
|
| 18 | c3cumA_
|
|
 |
72.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
|
|
|
| 19 | c2olsA_
|
|
 |
71.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
|
|
|
| 20 | c1kblA_
|
|
 |
71.4 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: pyruvate phosphate dikinase
|
|
|
|
| 21 | d1ma3a_ |
|
not modelled |
71.0 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
|
|
| 22 | c3afoB_ |
|
not modelled |
70.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:nadh kinase pos5;
PDBTitle: crystal structure of yeast nadh kinase complexed with nadh
|
|
|
| 23 | c2bg5C_ |
|
not modelled |
70.2 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme i-domain2 from the thermoanaerobacter tengcongensis pep: sugar3 phosphotransferase system (pts)
|
|
|
| 24 | d1xmta_ |
|
not modelled |
69.9 |
20 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 25 | d2hk6a1 |
|
not modelled |
65.9 |
14 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
|
|
| 26 | c3l4eA_ |
|
not modelled |
65.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized peptidase lmo0363;
PDBTitle: 1.5a crystal structure of a putative peptidase e protein from listeria2 monocytogenes egd-e
|
|
|
| 27 | c4e38A_ |
|
not modelled |
65.1 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 28 | c3mw8A_ |
|
not modelled |
65.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
|
|
| 29 | d1fyea_ |
|
not modelled |
64.4 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Aspartyl dipeptidase PepE |
|
|
| 30 | c5i0cA_ |
|
not modelled |
64.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein yjdj;
PDBTitle: crystal structure of predicted acyltransferase yjdj with acyl-coa n-2 acyltransferase domain from escherichia coli str. k-12
|
|
|
| 31 | c6a4tB_ |
|
not modelled |
62.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase e;
PDBTitle: crystal structure of peptidase e from deinococcus radiodurans r1
|
|
|
| 32 | c6q7jB_ |
|
not modelled |
61.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:exo-1,4-beta-xylosidase xlnd;
PDBTitle: gh3 exo-beta-xylosidase (xlnd) in complex with xylobiose aziridine2 activity based probe
|
|
|
| 33 | c3fg9B_ |
|
not modelled |
61.8 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein of universal stress protein uspa family;
PDBTitle: the crystal structure of an universal stress protein uspa2 family protein from lactobacillus plantarum wcfs1
|
|
|
| 34 | c3zdrA_ |
|
not modelled |
61.2 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase domain of the bifunctional
PDBTitle: structure of the alcohol dehydrogenase (adh) domain of a2 bifunctional adhe dehydrogenase from geobacillus3 thermoglucosidasius ncimb 11955
|
|
|
| 35 | c5ereA_ |
|
not modelled |
60.8 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: extracellular ligand binding receptor from desulfohalobium retbaense2 dsm5692
|
|
|
| 36 | c4maaA_ |
|
not modelled |
60.7 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:putative branched-chain amino acid abc transporter,
PDBTitle: the crystal structure of amino acid abc transporter substrate-binding2 protein from pseudomonas fluorescens pf-5
|
|
|
| 37 | c5vtoA_ |
|
not modelled |
60.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:blasticidin m;
PDBTitle: solution structure of blsm
|
|
|
| 38 | c4qccA_ |
|
not modelled |
59.9 |
16 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 39 | d1kbla1 |
|
not modelled |
58.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
|
|
| 40 | d1qo0a_ |
|
not modelled |
58.4 |
17 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 41 | c4evsA_ |
|
not modelled |
58.2 |
7 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter subunit, substrate-binding
PDBTitle: crystal structure of abc transporter from r. palustris - solute2 binding protein (rpa0985) in complex with 4-hydroxybenzoate
|
|
|
| 42 | c1jzdA_ |
|
not modelled |
55.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: dsbc-dsbdalpha complex
|
|
|
| 43 | d2d59a1 |
|
not modelled |
55.8 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 44 | c4yt2A_ |
|
not modelled |
55.6 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmd ii from methanocaldococcus jannaschii
|
|
|
| 45 | c6huxA_ |
|
not modelled |
54.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmdii from methanocaldococcus jannaschii reconstitued with fe-2 guanylylpyridinol (fegp) cofactor and co-crystallized with methenyl-3 tetrahydromethanopterin at 2.5 a resolution
|
|
|
| 46 | c3pkiF_ |
|
not modelled |
54.8 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
|
|
| 47 | c3w6uA_ |
|
not modelled |
54.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, nad-binding protein;
PDBTitle: crystal structure of nadp bound l-serine 3-dehydrogenase from2 hyperthermophilic archaeon pyrobaculum calidifontis
|
|
|
| 48 | c2iz6A_ |
|
not modelled |
54.5 |
18 |
PDB header:metal transport
Chain: A: PDB Molecule:molybdenum cofactor carrier protein;
PDBTitle: structure of the chlamydomonas rheinhardtii moco carrier2 protein
|
|
|
| 49 | c3ac0B_ |
|
not modelled |
54.4 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase i;
PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
|
|
|
| 50 | c5mrwF_ |
|
not modelled |
54.3 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:potassium-transporting atpase atp-binding subunit;
PDBTitle: structure of the kdpfabc complex
|
|
|
| 51 | c4mlcA_ |
|
not modelled |
54.1 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: abc transporter substrate-binding protein fromdesulfitobacterium2 hafniense
|
|
|
| 52 | c4dllB_ |
|
not modelled |
53.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-hydroxy-3-oxopropionate reductase;
PDBTitle: crystal structure of a 2-hydroxy-3-oxopropionate reductase from2 polaromonas sp. js666
|
|
|
| 53 | c5i01B_ |
|
not modelled |
53.4 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of phosphoheptose isomerase gmha from neisseria gonorrhoeae
|
|
|
| 54 | d2g17a1 |
|
not modelled |
53.2 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 55 | c1gqqA_ |
|
not modelled |
53.1 |
22 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
|
|
|
| 56 | c3snrA_ |
|
not modelled |
52.0 |
21 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
|
|
| 57 | c4n03A_ |
|
not modelled |
51.8 |
5 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type branched-chain amino acid transport systems
PDBTitle: fatty acid abc transporter substrate-binding protein from2 thermomonospora curvata
|
|
|
| 58 | d1yq2a5 |
|
not modelled |
51.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 59 | d2jfga1 |
|
not modelled |
50.8 |
17 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 60 | d1h6za1 |
|
not modelled |
50.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
|
|
| 61 | d1yc5a1 |
|
not modelled |
50.7 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
|
|
| 62 | d1r57a_ |
|
not modelled |
50.5 |
31 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 63 | c3h6hB_ |
|
not modelled |
50.3 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:glutamate receptor, ionotropic kainate 2;
PDBTitle: crystal structure of the glur6 amino terminal domain dimer assembly2 mpd form
|
|
|
| 64 | c1j6uA_ |
|
not modelled |
50.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
|
|
|
| 65 | d1vbga1 |
|
not modelled |
49.8 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
|
|
| 66 | c6cauA_ |
|
not modelled |
49.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
|
|
|
| 67 | c3i09A_ |
|
not modelled |
49.6 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic branched-chain amino acid-binding protein;
PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
|
|
|
| 68 | c4evrA_ |
|
not modelled |
49.5 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter subunit, substrate-binding
PDBTitle: crystal structure of abc transporter from r. palustris - solute2 binding protein (rpa0668) in complex with benzoate
|
|
|
| 69 | c2hroA_ |
|
not modelled |
49.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
|
|
| 70 | c3s40C_ |
|
not modelled |
49.1 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
|
|
| 71 | c1t3bA_ |
|
not modelled |
48.9 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: x-ray structure of dsbc from haemophilus influenzae
|
|
|
| 72 | d2j13a1 |
|
not modelled |
48.7 |
11 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
| 73 | c4gnrA_ |
|
not modelled |
48.3 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter substrate-binding protein-branched chain
PDBTitle: 1.0 angstrom resolution crystal structure of the branched-chain amino2 acid transporter substrate binding protein livj from streptococcus3 pneumoniae str. canada mdr_19a in complex with isoleucine
|
|
|
| 74 | c5f56A_ |
|
not modelled |
48.2 |
15 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:single-stranded-dna-specific exonuclease;
PDBTitle: structure of recj complexed with dna and ssb-ct
|
|
|
| 75 | c6hpdA_ |
|
not modelled |
48.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase (gh2);
PDBTitle: the structure of a beta-glucuronidase from glycoside hydrolase family2 2
|
|
|
| 76 | c3zg6A_ |
|
not modelled |
47.8 |
19 |
PDB header:hydrolase/inhibitor
Chain: A: PDB Molecule:nad-dependent protein deacetylase sirtuin-6;
PDBTitle: the novel de-long chain fatty acid function of human sirt6
|
|
|
| 77 | c3ej6D_ |
|
not modelled |
47.5 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:catalase-3;
PDBTitle: neurospora crassa catalase-3 crystal structure
|
|
|
| 78 | c4i3gB_ |
|
not modelled |
47.5 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of desr, a beta-glucosidase from streptomyces2 venezuelae in complex with d-glucose.
|
|
|
| 79 | c5g6sD_ |
|
not modelled |
47.2 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:imine reductase;
PDBTitle: imine reductase from aspergillus oryzae in complex with nadp(h) and2 (r)-rasagiline
|
|
|
| 80 | c3sg0A_ |
|
not modelled |
46.6 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
|
|
|
| 81 | c4npbA_ |
|
not modelled |
46.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:protein disulfide isomerase ii;
PDBTitle: the crystal structure of thiol:disulfide interchange protein dsbc from2 yersinia pestis co92
|
|
|
| 82 | d2je8a5 |
|
not modelled |
45.7 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 83 | c3eafA_ |
|
not modelled |
44.0 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein;
PDBTitle: crystal structure of abc transporter, substrate binding protein2 aeropyrum pernix
|
|
|
| 84 | c2hwgA_ |
|
not modelled |
43.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
|
|
|
| 85 | d1uc8a1 |
|
not modelled |
43.8 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Lysine biosynthesis enzyme LysX, N-terminal domain |
|
|
| 86 | c4l1gB_ |
|
not modelled |
43.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidoglycan n-acetylglucosamine deacetylase;
PDBTitle: crystal structure of the bc1960 peptidoglycan n-acetylglucosamine2 deacetylase from bacillus cereus
|
|
|
| 87 | d1w5fa1 |
|
not modelled |
43.0 |
14 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 88 | d7reqa2 |
|
not modelled |
42.5 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 89 | c3n0xA_ |
|
not modelled |
42.3 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:possible substrate binding protein of abc transporter
PDBTitle: crystal structure of an abc-type branched-chain amino acid transporter2 (rpa4397) from rhodopseudomonas palustris cga009 at 1.50 a resolution
|
|
|
| 90 | c5nymA_ |
|
not modelled |
42.3 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin-like protein 2.1;
PDBTitle: crystal structure of the atypical poplar thioredoxin-like2.1 in2 reduced state
|
|
|
| 91 | d1t3ba1 |
|
not modelled |
42.3 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbC/DsbG C-terminal domain-like |
|
|
| 92 | c4f11A_ |
|
not modelled |
42.2 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:gamma-aminobutyric acid type b receptor subunit 2;
PDBTitle: crystal structure of the extracellular domain of human gaba(b)2 receptor gbr2
|
|
|
| 93 | c1vbhA_ |
|
not modelled |
42.1 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
|
|
| 94 | d1vhca_ |
|
not modelled |
42.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 95 | d1k0ma2 |
|
not modelled |
41.9 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
|
|
| 96 | c3mn1B_ |
|
not modelled |
41.5 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable yrbi family phosphatase;
PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
|
|
|
| 97 | d2hrca1 |
|
not modelled |
41.4 |
16 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
|
|
| 98 | c4umwA_ |
|
not modelled |
41.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc-transporting atpase;
PDBTitle: crystal structure of a zinc-transporting pib-type atpase in2 e2.pi state
|
|
|
| 99 | d1p3da1 |
|
not modelled |
41.0 |
24 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 100 | c3b8cB_ |
|
not modelled |
40.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:atpase 2, plasma membrane-type;
PDBTitle: crystal structure of a plasma membrane proton pump
|
|
|
| 101 | c4kv7A_ |
|
not modelled |
40.5 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:probable leucine/isoleucine/valine-binding protein;
PDBTitle: the crystal structure of a possible leucine/isoleucine/valine-binding2 protein from rhodopirellula baltica sh 1
|
|
|
| 102 | c3fwzA_ |
|
not modelled |
40.2 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
|
|
| 103 | d1vlja_ |
|
not modelled |
40.1 |
13 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 104 | d2dlxa1 |
|
not modelled |
39.9 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:UAS domain |
|
|
| 105 | c5kuhB_ |
|
not modelled |
39.7 |
11 |
PDB header:signaling protein
Chain: B: PDB Molecule:glutamate receptor ionotropic, kainate 2;
PDBTitle: gluk2em with ly466195
|
|
|
| 106 | c6qkzA_ |
|
not modelled |
39.5 |
8 |
PDB header:membrane protein
Chain: A: PDB Molecule:glua1;
PDBTitle: full length glua1/2-gamma8 complex
|
|
|
| 107 | c3cmgA_ |
|
not modelled |
39.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
|
|
| 108 | c2r8zC_ |
|
not modelled |
39.0 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
|
|
| 109 | d1s5pa_ |
|
not modelled |
38.7 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
|
|
| 110 | c2vpiA_ |
|
not modelled |
38.7 |
3 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
|
|
| 111 | c1v57A_ |
|
not modelled |
38.7 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbg;
PDBTitle: crystal structure of the disulfide bond isomerase dsbg
|
|
|
| 112 | d1s1pa_ |
|
not modelled |
38.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
|
|
| 113 | c4ru0B_ |
|
not modelled |
38.5 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc transporter,
PDBTitle: the crystal structure of abc transporter permease from pseudomonas2 fluorescens group
|
|
|
| 114 | c3f4cA_ |
|
not modelled |
38.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:organophosphorus hydrolase;
PDBTitle: crystal structure of organophosphorus hydrolase from geobacillus2 stearothermophilus strain 10, with glycerol bound
|
|
|
| 115 | d1iuka_ |
|
not modelled |
38.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 116 | c3p9zA_ |
|
not modelled |
38.2 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
|
|
| 117 | c2mtbA_ |
|
not modelled |
37.8 |
5 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin-2;
PDBTitle: solution structure of apo_fldb
|
|
|
| 118 | c4nqrA_ |
|
not modelled |
37.4 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid/amide abc transporter substrate-binding protein,
PDBTitle: the crystal structure of a solute-binding protein (n280d mutant) from2 anabaena variabilis atcc 29413 in complex with alanine
|
|
|
| 119 | d1ofua1 |
|
not modelled |
37.3 |
16 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 120 | d1jhfa1 |
|
not modelled |
37.3 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
|
|