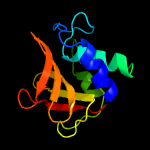
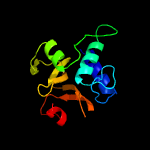
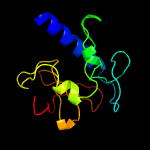
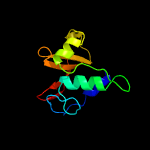
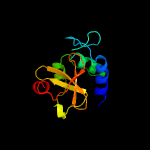
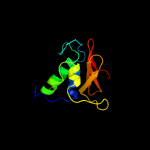
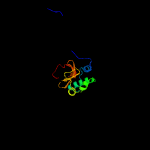
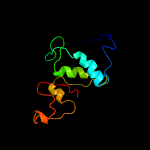
1 c2xivA_
99.9
21
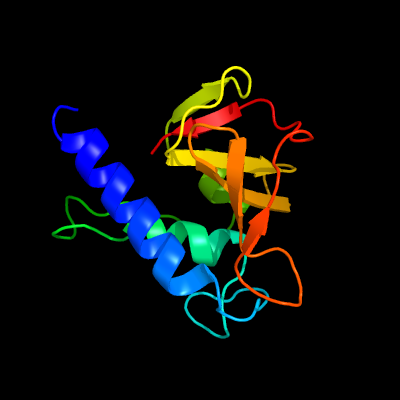
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of mycobacterium2 tuberculosis
2 c6biqA_
99.9
17
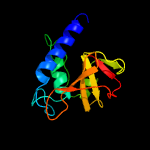
PDB header: hydrolaseChain: A: PDB Molecule: clan ca, family c40, nlpc/p60 superfamily cysteinePDBTitle: structure of nlpc2 from trichomonas vaginalis
3 c3pbiA_
99.9
21
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
4 c6b8cA_
99.9
28
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60;PDBTitle: crystal structure of nlpc/p60 domain of peptidoglycan hydrolase saga
5 c2fg0B_
99.9
17
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
6 c3gt2A_
99.9
21
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium paratuberculosis2 antigen map1272c
7 d2evra2
99.9
20
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P60
8 c4fdyA_
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: similar to lipoprotein, nlp/p60 family;PDBTitle: crystal structure of a similar to lipoprotein, nlp/p60 family2 (sav0400) from staphylococcus aureus subsp. aureus mu50 at 2.23 a3 resolution
9 c3npfB_
99.9
15
PDB header: hydrolaseChain: B: PDB Molecule: putative dipeptidyl-peptidase vi;PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
10 c3i86A_
99.9
22
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
11 c2k1gA_
99.9
16
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
12 c4hpeA_
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: putative cell wall hydrolase tn916-like,ctn1-orf17;PDBTitle: crystal structure of a putative cell wall hydrolase (cd630_03720) from2 clostridium difficile 630 at 2.38 a resolution
13 c3h41A_
99.8
19
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
14 c4xcmB_
99.7
23
PDB header: hydrolaseChain: B: PDB Molecule: cell wall-binding endopeptidase-related protein;PDBTitle: crystal structure of the putative nlpc/p60 d,l endopeptidase from t.2 thermophilus
15 c3m1uB_
99.5
17
PDB header: hydrolaseChain: B: PDB Molecule: putative gamma-d-glutamyl-l-diamino acid endopeptidase;PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
16 d2if6a1
95.4
22
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like
17 c2p1gA_
94.3
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative xylanase;PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
18 c5udmA_
93.1
21
PDB header: hydrolaseChain: A: PDB Molecule: phage-associated cell wall hydrolase;PDBTitle: phage-associated cell wall hydrolase plypy from streptococcus2 pyogenes, space group p6522
19 c2lktA_
88.3
28
PDB header: hydrolaseChain: A: PDB Molecule: retinoic acid receptor responder protein 3;PDBTitle: solution structure of n-terminal domain of human tig3 in 2 m urea
20 c2kytA_
86.8
25
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution structure of the h-rev107 n-terminal domain
21 c2im9A_
not modelled
86.7
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
22 d2im9a1
not modelled
86.7
28
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Lpg0564-like
23 c4h4jA_
not modelled
86.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a n-acetylmuramoyl-l-alanine amidase2 (bacuni_02947) from bacteroides uniformis atcc 8492 at 1.15 a3 resolution
24 c4pa5A_
not modelled
80.1
15
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: tgl - a bacterial spore coat transglutaminase - cystamine complex
25 c3kw0D_
not modelled
76.2
23
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
26 c4f0wA_
not modelled
71.7
18
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of type effector tse1 c30a mutant from pseudomonas2 aeruginousa
27 c4f4mA_
not modelled
70.9
18
PDB header: hydrolase regulatorChain: A: PDB Molecule: papain peptidoglycan amidase effector tse1;PDBTitle: structure of the type vi peptidoglycan amidase effector tse1 (c30a)2 from pseudomonas aeruginosa
28 c2lrjA_
not modelled
69.2
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: staphyloxanthin biosynthesis protein, putative;PDBTitle: nmr solution structure of staphyloxanthin biosynthesis protein
29 c2k3aA_
not modelled
64.8
22
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
30 c5t1qB_
not modelled
61.7
22
PDB header: hydrolaseChain: B: PDB Molecule: n-acetylmuramoyl-l-alanine amidase domain-containingPDBTitle: 2.15 angstrom crystal structure of n-acetylmuramoyl-l-alanine amidase2 from staphylococcus aureus.
31 c4hzbA_
not modelled
40.6
22
PDB header: hydrolaseChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: crystal structure of the type vi semet effector-immunity complex tae3-2 tai3 from ralstonia pickettii
32 c4hfkB_
not modelled
37.5
14
PDB header: hydrolaseChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the type vi effector-immunity complex tae4-tai42 from enterobacter cloacae
33 c4olkB_
not modelled
31.7
22
PDB header: hydrolaseChain: B: PDB Molecule: endolysin;PDBTitle: the chap domain of lysgh15
34 d1f35a_
not modelled
19.2
17
Fold: Olfactory marker proteinSuperfamily: Olfactory marker proteinFamily: Olfactory marker protein
35 c6avaA_
not modelled
13.0
36
PDB header: toxinChain: A: PDB Molecule: insectotoxin-i1;PDBTitle: exploring cystine dense peptide space to open a unique molecular2 toolbox
36 c4b6iD_
not modelled
12.6
17
PDB header: signaling proteinChain: D: PDB Molecule: sma2266;PDBTitle: crystal structure rap2b (sma2266) from serratia marcescens
37 c3cjiC_
not modelled
10.0
27
PDB header: virusChain: C: PDB Molecule: polyprotein;PDBTitle: structure of seneca valley virus-001
38 d1iyca_
not modelled
6.5
44
Fold: Invertebrate chitin-binding proteinsSuperfamily: Invertebrate chitin-binding proteinsFamily: Antifungal peptide scarabaecin
39 c3zibD_
not modelled
6.5
21
PDB header: protein bindingChain: D: PDB Molecule: rap2a sma2265;PDBTitle: rap2a protein (sma2265) from serratia marcescens
40 c2vpmB_
not modelled
6.2
24
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
41 d2io8a2
not modelled
5.8
39
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: CHAP domain
42 c2ls1A_
not modelled
5.7
56
PDB header: antimicrobial proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of sviceucin, an antibacterial type i lasso peptide from2 streptomyces sviceus
43 c5fx8U_
not modelled
5.7
45
PDB header: oxidoreductaseChain: U: PDB Molecule: zonadhesin;PDBTitle: complete structure of manganese lipoxygenase of gaeumannomyces2 graminis and partial structure of zonadhesin of komagataella3 pastoris