1 c3kw0D_
100.0
17
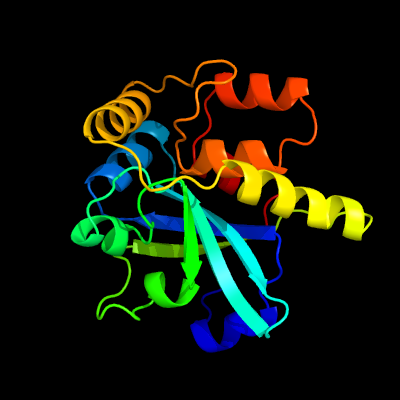
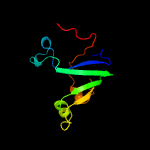
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
2 d2if6a1
99.9
25
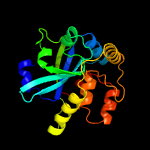
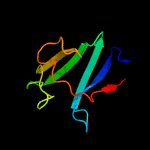
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like
3 c2p1gA_
97.0
20
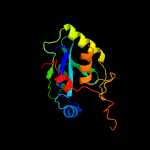
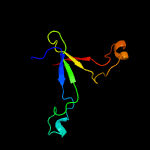
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative xylanase;PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
4 d2evra2
96.3
20
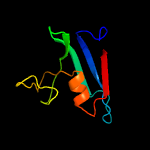
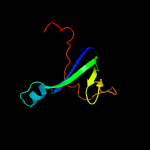
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P60
5 c2fg0B_
95.7
24
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
6 c6biqA_
95.3
29
PDB header: hydrolaseChain: A: PDB Molecule: clan ca, family c40, nlpc/p60 superfamily cysteinePDBTitle: structure of nlpc2 from trichomonas vaginalis
7 c3npfB_
94.3
19
PDB header: hydrolaseChain: B: PDB Molecule: putative dipeptidyl-peptidase vi;PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
8 c2k1gA_
94.3
39
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
9 c4h4jA_
93.7
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a n-acetylmuramoyl-l-alanine amidase2 (bacuni_02947) from bacteroides uniformis atcc 8492 at 1.15 a3 resolution
10 c6b8cA_
93.5
20
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60;PDBTitle: crystal structure of nlpc/p60 domain of peptidoglycan hydrolase saga
11 c3h41A_
93.2
24
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
12 c3pbiA_
92.6
20
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
13 c2xivA_
92.1
22
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of mycobacterium2 tuberculosis
14 c2im9A_
91.7
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
15 d2im9a1
91.7
11
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Lpg0564-like
16 c4hpeA_
91.6
8
PDB header: hydrolaseChain: A: PDB Molecule: putative cell wall hydrolase tn916-like,ctn1-orf17;PDBTitle: crystal structure of a putative cell wall hydrolase (cd630_03720) from2 clostridium difficile 630 at 2.38 a resolution
17 c4fdyA_
90.5
10
PDB header: hydrolaseChain: A: PDB Molecule: similar to lipoprotein, nlp/p60 family;PDBTitle: crystal structure of a similar to lipoprotein, nlp/p60 family2 (sav0400) from staphylococcus aureus subsp. aureus mu50 at 2.23 a3 resolution
18 c3gt2A_
90.3
13
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium paratuberculosis2 antigen map1272c
19 d2io8a2
87.5
12
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: CHAP domain
20 c4xcmB_
86.6
24
PDB header: hydrolaseChain: B: PDB Molecule: cell wall-binding endopeptidase-related protein;PDBTitle: crystal structure of the putative nlpc/p60 d,l endopeptidase from t.2 thermophilus
21 c4olkB_
not modelled
85.5
16
PDB header: hydrolaseChain: B: PDB Molecule: endolysin;PDBTitle: the chap domain of lysgh15
22 c3i86A_
not modelled
76.5
18
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
23 c2lrjA_
not modelled
68.0
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: staphyloxanthin biosynthesis protein, putative;PDBTitle: nmr solution structure of staphyloxanthin biosynthesis protein
24 c3mvnA_
not modelled
60.5
17
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-medo-PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamyl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
25 d2jfga2
not modelled
57.6
15
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
26 c2vpmB_
not modelled
54.8
24
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
27 c2am1A_
not modelled
48.2
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine-d-glutamyl-lysine-d-alanyl-d-PDBTitle: sp protein ligand 1
28 c2lktA_
not modelled
44.4
18
PDB header: hydrolaseChain: A: PDB Molecule: retinoic acid receptor responder protein 3;PDBTitle: solution structure of n-terminal domain of human tig3 in 2 m urea
29 c4bubA_
not modelled
42.1
30
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--ld-lysinePDBTitle: crystal structure of mure ligase from thermotoga maritima2 in complex with adp
30 c2f00A_
not modelled
42.0
17
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
31 c2wtzC_
not modelled
41.7
30
PDB header: ligaseChain: C: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate-PDBTitle: mure ligase of mycobacterium tuberculosis
32 c1e8cB_
not modelled
41.3
20
PDB header: ligaseChain: B: PDB Molecule: udp-n-acetylmuramoylalanyl-d-glutamate--2,6-diaminopimelatePDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide synthetase from2 e. coli
33 c3uagA_
not modelled
41.3
18
PDB header: ligaseChain: A: PDB Molecule: protein (udp-n-acetylmuramoyl-l-alanine:d-PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
34 c3lk7A_
not modelled
41.1
22
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine--d-glutamate ligase;PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-glutamate2 (murd) ligase from streptococcus agalactiae to 1.5a
35 c5ibzD_
not modelled
38.5
32
PDB header: lyaseChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a novel cyclase (pfam04199).
36 c2k3aA_
not modelled
37.8
12
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
37 d1ofla_
not modelled
33.6
21
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Chondroitinase B
38 c4eyzB_
not modelled
33.2
18
PDB header: hydrolaseChain: B: PDB Molecule: cellulosome-related protein module from ruminococcusPDBTitle: crystal structure of an uncommon cellulosome-related protein module2 from ruminococcus flavefaciens that resembles papain-like cysteine3 peptidases
39 c5udmA_
not modelled
32.8
24
PDB header: hydrolaseChain: A: PDB Molecule: phage-associated cell wall hydrolase;PDBTitle: phage-associated cell wall hydrolase plypy from streptococcus2 pyogenes, space group p6522
40 c4c13A_
not modelled
31.5
15
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--l-lysine ligase;PDBTitle: x-ray crystal structure of staphylococcus aureus mure with udp-murnac-2 ala-glu-lys
41 c2xjaD_
not modelled
31.3
30
PDB header: ligaseChain: D: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--2,6-PDBTitle: structure of mure from m.tuberculosis with dipeptide and adp
42 c4ba6A_
not modelled
30.4
36
PDB header: carbohydrate-binding proteinChain: A: PDB Molecule: endoglucanase cel5a;PDBTitle: high resolution structure of the c-terminal family 65 carbohydrate2 binding module (cbm65b) of endoglucanase cel5a from eubacterium3 cellulosolvens
43 c4bucA_
not modelled
29.6
22
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine--d-glutamate ligase;PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
44 c3m1uB_
not modelled
29.1
10
PDB header: hydrolaseChain: B: PDB Molecule: putative gamma-d-glutamyl-l-diamino acid endopeptidase;PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
45 d2v4ja2
not modelled
27.1
47
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like
46 d1p3da2
not modelled
27.1
24
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
47 d1e8ca2
not modelled
25.8
25
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
48 c3hn7A_
not modelled
23.6
14
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
49 c1dbgA_
not modelled
23.0
21
PDB header: lyaseChain: A: PDB Molecule: chondroitinase b;PDBTitle: crystal structure of chondroitinase b
50 c2ioaA_
not modelled
22.8
14
PDB header: ligase, hydrolaseChain: A: PDB Molecule: bifunctional glutathionylspermidinePDBTitle: e. coli bifunctional glutathionylspermidine2 synthetase/amidase incomplex with mg2+ and adp and3 phosphinate inhibitor
51 d1y88a2
not modelled
21.9
31
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: MRR-like
52 c4lt5A_
not modelled
20.1
33
PDB header: oxidoreductase/dnaChain: A: PDB Molecule: naegleria tet-like dioxygenase;PDBTitle: structure of a naegleria tet-like dioxygenase in complex with 5-2 methylcytosine dna
53 c2eb0B_
not modelled
18.9
15
PDB header: hydrolaseChain: B: PDB Molecule: manganese-dependent inorganic pyrophosphatase;PDBTitle: crystal structure of methanococcus jannaschii putative family ii2 inorganic pyrophosphatase
54 c1gqqA_
not modelled
18.4
24
PDB header: cell wall biosynthesisChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
55 c2kytA_
not modelled
18.4
17
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution structure of the h-rev107 n-terminal domain
56 c4pyzA_
not modelled
17.8
30
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase 7;PDBTitle: crystal structure of the first two ubl domains of deubiquitylase usp7
57 c6odmK_
not modelled
16.0
22
PDB header: viral proteinChain: K: PDB Molecule: capsid vertex component 2;PDBTitle: herpes simplex virus type 1 (hsv-1) portal vertex-adjacent2 capsid/catc, asymmetric unit
58 c3vsvD_
not modelled
14.9
16
PDB header: hydrolaseChain: D: PDB Molecule: xylosidase;PDBTitle: the complex structure of xylc with xylose
59 d1k20a_
not modelled
14.3
14
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)
60 c5t1qB_
not modelled
13.9
16
PDB header: hydrolaseChain: B: PDB Molecule: n-acetylmuramoyl-l-alanine amidase domain-containingPDBTitle: 2.15 angstrom crystal structure of n-acetylmuramoyl-l-alanine amidase2 from staphylococcus aureus.
61 d2ahua2
not modelled
13.6
24
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
62 c1w78A_
not modelled
12.9
17
PDB header: synthaseChain: A: PDB Molecule: folc bifunctional protein;PDBTitle: e.coli folc in complex with dhpp and adp
63 d1wpna_
not modelled
12.9
21
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)
64 c2fphX_
not modelled
12.9
26
PDB header: dna binding proteinChain: X: PDB Molecule: ylmh;PDBTitle: cell division protein ylmh from streptococcus pneumoniae
65 d1s04a_
not modelled
12.6
25
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain
66 d1d02a_
not modelled
12.6
26
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease MunI
67 c4rpaA_
not modelled
12.3
21
PDB header: hydrolaseChain: A: PDB Molecule: probable manganese-dependent inorganic pyrophosphatase;PDBTitle: crystal structure of inorganic pyrophosphatase from staphylococcus2 aureus in complex with mn2+
68 c3a9lB_
not modelled
12.1
33
PDB header: hydrolaseChain: B: PDB Molecule: poly-gamma-glutamate hydrolase;PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
69 d1uw4a_
not modelled
11.4
27
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Smg-4/UPF3
70 d1i74a_
not modelled
11.2
29
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)
71 d1ltqa1
not modelled
11.2
20
Fold: HAD-likeSuperfamily: HAD-likeFamily: phosphatase domain of polynucleotide kinase
72 c6bs5B_
not modelled
10.7
25
PDB header: unknown functionChain: B: PDB Molecule: anion transporter;PDBTitle: crystal structure of amp-pnp-bound bacterial get3-like a and b in2 mycobacterium tuberculosis
73 c3j6vQ_
not modelled
10.4
25
PDB header: ribosomeChain: Q: PDB Molecule: 28s ribosomal protein s17, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
74 c4yooA_
not modelled
10.4
20
PDB header: transcriptionChain: A: PDB Molecule: retinoblastoma-like protein 1,retinoblastoma-like proteinPDBTitle: p107 pocket domain in complex with lin52 p29a peptide
75 d1o57a1
not modelled
10.3
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: N-terminal domain of Bacillus PurR
76 c5dniB_
not modelled
10.3
17
PDB header: lyaseChain: B: PDB Molecule: putative l(+)-tartrate dehydratase subunit beta;PDBTitle: crystal structure of methanocaldococcus jannaschii fumarate hydratase2 beta subunit
77 c6hwhX_
not modelled
10.2
21
PDB header: electron transportChain: X: PDB Molecule: cytochrome c oxidase polypeptide 4;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
78 d2a1xa1
not modelled
10.0
16
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like
79 d1zj8a1
not modelled
8.6
16
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
80 c4j0nA_
not modelled
8.6
25
PDB header: hydrolaseChain: A: PDB Molecule: isatin hydrolase b;PDBTitle: crystal structure of a manganese dependent isatin hydrolase
81 c5ykaA_
not modelled
8.2
27
PDB header: biosynthetic proteinChain: A: PDB Molecule: uncharacterized protein kdoo;PDBTitle: crystal structure of the kdo hydroxylase kdoo, a non-heme fe(ii)2 alphaketoglutarate dependent dioxygenase in complex with cobalt(ii)
82 c4m1bA_
not modelled
8.1
17
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: structural determination of ba0150, a polysaccharide deacetylase from2 bacillus anthracis
83 d2g40a1
not modelled
8.0
23
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: YkgG-like
84 c2g40A_
not modelled
8.0
23
PDB header: unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a duf162 family protein (dr_1909) from2 deinococcus radiodurans at 1.70 a resolution
85 d2hawa1
not modelled
8.0
21
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)
86 c1j6uA_
not modelled
7.8
14
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
87 d1e0ta1
not modelled
7.7
11
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain
88 d1g03a_
not modelled
7.6
33
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain
89 d2g50a1
not modelled
7.6
11
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain
90 d2uubp1
not modelled
7.6
12
Fold: Ribosomal protein S16Superfamily: Ribosomal protein S16Family: Ribosomal protein S16
91 d1d8ja_
not modelled
7.4
47
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: The central core domain of TFIIE beta
92 d1wu2a2
not modelled
7.3
16
Fold: MoeA N-terminal region -likeSuperfamily: MoeA N-terminal region -likeFamily: MoeA N-terminal region -like
93 c3c7xA_
not modelled
7.3
23
PDB header: hydrolaseChain: A: PDB Molecule: matrix metalloproteinase-14;PDBTitle: hemopexin-like domain of matrix metalloproteinase 14
94 c5nmpF_
not modelled
7.1
26
PDB header: hydrolaseChain: F: PDB Molecule: isatin hydrolase;PDBTitle: isatin hydrolase a (iha) from ralstonia solanacearum
95 d1a3xa1
not modelled
7.1
5
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain
96 c5lz6B_
not modelled
7.1
29
PDB header: antiviral proteinChain: B: PDB Molecule: 3a;PDBTitle: crystal structure of human acbd3 gold domain in complex with 3a2 protein of aichivirus b
97 c4akrC_
not modelled
7.1
26
PDB header: actin-binding proteinChain: C: PDB Molecule: f-actin-capping protein subunit alpha;PDBTitle: crystal structure of the cytoplasmic actin capping protein2 cap32_34 from dictyostelium discoideum
98 c5onkA_
not modelled
7.1
16
PDB header: hydrolaseChain: A: PDB Molecule: yndl;PDBTitle: native yndl
99 d1zgha2
not modelled
6.9
21
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase