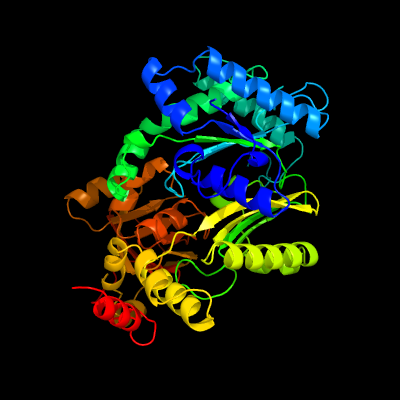
| 1 | c1kblA_
|
|
 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: pyruvate phosphate dikinase
|
|
|
|
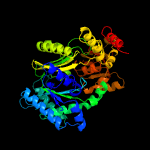
| 2 | c1h6zA_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
|
|
|
|
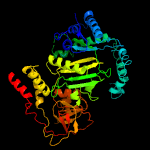
| 3 | c1vbhA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
|
|
|
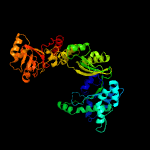
| 4 | c2olsA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
|
|
|
| 5 | c5fbtA_
|
|
 |
100.0 |
32 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: crystal structure of rifampin phosphotransferase rph-lm from listeria2 monocytogenes in complex with rifampin
|
|
|
|
| 6 | d1kbla3
|
|
 |
100.0 |
40 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Pyruvate phosphate dikinase, N-terminal domain |
|
|
|
| 7 | d1vbga3
|
|
 |
100.0 |
41 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Pyruvate phosphate dikinase, N-terminal domain |
|
|
|
| 8 | d1h6za3
|
|
 |
100.0 |
37 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Pyruvate phosphate dikinase, N-terminal domain |
|
|
|
| 9 | d1kbla2
|
|
 |
100.0 |
44 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
|
| 10 | d1vbga2
|
|
 |
100.0 |
40 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
|
| 11 | d1h6za2
|
|
 |
100.0 |
41 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
|
| 12 | d1zyma2
|
|
 |
99.9 |
25 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system |
|
|
|
| 13 | c3t07D_
|
|
 |
99.9 |
23 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
|
|
|
|
| 14 | c2e28A_
|
|
 |
99.9 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
|
|
|
|
| 15 | c5woyA_
|
|
 |
99.9 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: nmr solution structure of enzyme i (neit) protein using two 4d-spectra
|
|
|
|
| 16 | c2hwgA_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
|
|
|
|
| 17 | c1ezaA_
|
|
 |
99.9 |
21 |
PDB header:phosphotransferase
Chain: A: PDB Molecule:enzyme i;
PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
|
|
|
|
| 18 | c5t1oB_
|
|
 |
99.8 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase ptsp;
PDBTitle: solution-state nmr and saxs structural ensemble of npr (1-85) in2 complex with ein-ntr (170-424)
|
|
|
|
| 19 | c2hroA_
|
|
 |
99.4 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
|
|
|
| 20 | d2hi6a1
|
|
 |
96.4 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:AF0055-like |
|
|
|
| 21 | c5ym0A_ |
|
not modelled |
94.3 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the crystal structure of dhad
|
|
|
| 22 | d2gp4a1 |
|
not modelled |
92.7 |
26 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
|
|
| 23 | c5oynB_ |
|
not modelled |
90.4 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:dehydratase, ilvd/edd family;
PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
|
|
|
| 24 | c2gp4B_ |
|
not modelled |
89.8 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
| 25 | c2pkpA_ |
|
not modelled |
88.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:homoaconitase small subunit;
PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
|
|
|
| 26 | c2gp4A_ |
|
not modelled |
85.6 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
| 27 | d1v7la_ |
|
not modelled |
79.0 |
34 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:LeuD-like |
|
|
| 28 | c3vbaE_ |
|
not modelled |
76.9 |
18 |
PDB header:lyase
Chain: E: PDB Molecule:isopropylmalate/citramalate isomerase small subunit;
PDBTitle: crystal structure of methanogen 3-isopropylmalate isomerase small2 subunit
|
|
|
| 29 | c5ze4A_ |
|
not modelled |
75.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
|
|
|
| 30 | c5j84A_ |
|
not modelled |
73.7 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase;
PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
|
|
|
| 31 | d5easa1 |
|
not modelled |
67.9 |
20 |
Fold:alpha/alpha toroid
Superfamily:Terpenoid cyclases/Protein prenyltransferases
Family:Terpenoid cyclase N-terminal domain |
|
|
| 32 | d1l5ja2 |
|
not modelled |
67.6 |
31 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:LeuD-like |
|
|
| 33 | c1m6vE_ |
|
not modelled |
66.6 |
18 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
|
|
| 34 | c3ln7A_ |
|
not modelled |
63.5 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 pasteurella multocida
|
|
|
| 35 | d1uc8a2 |
|
not modelled |
62.1 |
24 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Lysine biosynthesis enzyme LysX ATP-binding domain |
|
|
| 36 | c1w2wJ_ |
|
not modelled |
57.8 |
22 |
PDB header:isomerase
Chain: J: PDB Molecule:5-methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-phosphate2 isomerase related to regulatory eif2b subunits
|
|
|
| 37 | c3wvqA_ |
|
not modelled |
56.3 |
11 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:pgm1;
PDBTitle: structure of atp grasp protein
|
|
|
| 38 | c2zkrf_ |
|
not modelled |
55.7 |
13 |
PDB header:ribosomal protein/rna
Chain: F: PDB Molecule:rna expansion segment es7 part iii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
|
|
|
| 39 | c3ln6A_ |
|
not modelled |
53.3 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 streptococcus agalactiae
|
|
|
| 40 | c3r23B_ |
|
not modelled |
52.8 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine--d-alanine ligase from bacillus2 anthracis
|
|
|
| 41 | c2yvkA_ |
|
not modelled |
50.4 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
|
|
| 42 | c2hcuA_ |
|
not modelled |
49.6 |
30 |
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus mutans
|
|
|
| 43 | d1kska3 |
|
not modelled |
42.2 |
39 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Pseudouridine synthase RsuA N-terminal domain |
|
|
| 44 | c6a34B_ |
|
not modelled |
41.5 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:putative methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate isomerase from2 pyrococcus horikoshii ot3 - form i
|
|
|
| 45 | c1hx9A_ |
|
not modelled |
41.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:5-epi-aristolochene synthase;
PDBTitle: crystal structure of teas w273s form 1
|
|
|
| 46 | c3iz5H_ |
|
not modelled |
40.0 |
10 |
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein l7a (l7ae);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
|
|
| 47 | d1iowa2 |
|
not modelled |
37.5 |
25 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
|
|
| 48 | c3u9sA_ |
|
not modelled |
36.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
|
|
| 49 | c4gaxA_ |
|
not modelled |
35.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:amorpha-4,11-diene synthase;
PDBTitle: crystal structure of an alpha-bisabolol synthase mutant
|
|
|
| 50 | c3g4dB_ |
|
not modelled |
33.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:(+)-delta-cadinene synthase isozyme xc1;
PDBTitle: crystal structure of (+)-delta-cadinene synthase from gossypium2 arboreum and evolutionary divergence of metal binding motifs for3 catalysis
|
|
|
| 51 | c2h1fB_ |
|
not modelled |
32.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:lipopolysaccharide heptosyltransferase-1;
PDBTitle: e. coli heptosyltransferase waac with adp
|
|
|
| 52 | c4a1eF_ |
|
not modelled |
32.1 |
10 |
PDB header:ribosome
Chain: F: PDB Molecule:rpl7a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
|
|
|
| 53 | d1n1ba1 |
|
not modelled |
31.5 |
16 |
Fold:alpha/alpha toroid
Superfamily:Terpenoid cyclases/Protein prenyltransferases
Family:Terpenoid cyclase N-terminal domain |
|
|
| 54 | c3j39G_ |
|
not modelled |
31.1 |
13 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l7a;
PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
|
|
|
| 55 | c1ehiB_ |
|
not modelled |
30.9 |
7 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine:d-lactate ligase;
PDBTitle: d-alanine:d-lactate ligase (lmddl2) of vancomycin-resistant2 leuconostoc mesenteroides
|
|
|
| 56 | c2zbtB_ |
|
not modelled |
30.7 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: crystal structure of pyridoxine biosynthesis protein from thermus2 thermophilus hb8
|
|
|
| 57 | c2rkbE_ |
|
not modelled |
30.7 |
37 |
PDB header:lyase
Chain: E: PDB Molecule:serine dehydratase-like;
PDBTitle: serine dehydratase like-1 from human cancer cells
|
|
|
| 58 | c5b04B_ |
|
not modelled |
30.7 |
15 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 59 | c3zf7x_ |
|
not modelled |
30.0 |
13 |
PDB header:ribosome
Chain: X: PDB Molecule:60s ribosomal protein l23a;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 60 | d1ehia2 |
|
not modelled |
30.0 |
5 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
|
|
| 61 | c3tovB_ |
|
not modelled |
29.5 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
|
|
| 62 | d1vkza3 |
|
not modelled |
28.9 |
9 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
|
|
| 63 | c3n6rK_ |
|
not modelled |
28.6 |
16 |
PDB header:ligase
Chain: K: PDB Molecule:propionyl-coa carboxylase, alpha subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
|
|
| 64 | c2hjwA_ |
|
not modelled |
28.6 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: crystal structure of the bc domain of acc2
|
|
|
| 65 | c5h80A_ |
|
not modelled |
28.5 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:carboxylase;
PDBTitle: biotin carboxylase domain of single-chain bacterial carboxylase
|
|
|
| 66 | c3femB_ |
|
not modelled |
28.5 |
17 |
PDB header:biosynthetic protein, transferase
Chain: B: PDB Molecule:pyridoxine biosynthesis protein snz1;
PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
|
|
|
| 67 | c4zhtB_ |
|
not modelled |
28.4 |
6 |
PDB header:isomerase
Chain: B: PDB Molecule:bifunctional udp-n-acetylglucosamine 2-epimerase/n-
PDBTitle: crystal structure of udp-glcnac 2-epimerase
|
|
|
| 68 | c2i80B_ |
|
not modelled |
28.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine-d-alanine ligase;
PDBTitle: allosteric inhibition of staphylococcus aureus d-alanine:d-alanine2 ligase revealed by crystallographic studies
|
|
|
| 69 | c5ig9H_ |
|
not modelled |
27.8 |
14 |
PDB header:ligase
Chain: H: PDB Molecule:atp grasp ligase;
PDBTitle: crystal structure of macrocyclase mdnc bound with precursor peptide2 mdna from microcystis aeruginosa mrc
|
|
|
| 70 | c5mlkA_ |
|
not modelled |
27.8 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: biotin dependent carboxylase acca3 dimer from mycobacterium2 tuberculosis (rv3285)
|
|
|
| 71 | c5dmxC_ |
|
not modelled |
27.5 |
24 |
PDB header:ligase
Chain: C: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine-d-alanine ligase from acinetobacter2 baumannii, space group p212121
|
|
|
| 72 | d1iwpg_ |
|
not modelled |
27.5 |
27 |
Fold:Open three-helical up-and-down bundle
Superfamily:Diol dehydratase, gamma subunit
Family:Diol dehydratase, gamma subunit |
|
|
| 73 | c3j61G_ |
|
not modelled |
26.9 |
10 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l8e;
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
|
|
| 74 | d1k4ia_ |
|
not modelled |
26.9 |
14 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
|
|
| 75 | c3tqtB_ |
|
not modelled |
26.9 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: structure of the d-alanine-d-alanine ligase from coxiella burnetii
|
|
|
| 76 | d2cqaa1 |
|
not modelled |
26.8 |
30 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TIP49 domain |
|
|
| 77 | c1uc5M_ |
|
not modelled |
26.4 |
24 |
PDB header:lyase
Chain: M: PDB Molecule:diol dehydrase gamma subunit;
PDBTitle: structure of diol dehydratase complexed with (r)-1,2-2 propanediol
|
|
|
| 78 | d1eexg_ |
|
not modelled |
26.4 |
24 |
Fold:Open three-helical up-and-down bundle
Superfamily:Diol dehydratase, gamma subunit
Family:Diol dehydratase, gamma subunit |
|
|
| 79 | c3izcH_ |
|
not modelled |
26.2 |
13 |
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein rpl8 (l7ae);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 80 | c5i47A_ |
|
not modelled |
26.2 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:rimk domain protein atp-grasp;
PDBTitle: crystal structure of rimk domain protein atp-grasp from sphaerobacter2 thermophilus dsm 20745
|
|
|
| 81 | d3etja3 |
|
not modelled |
26.0 |
17 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
|
|
| 82 | c3u9sE_ |
|
not modelled |
25.9 |
18 |
PDB header:ligase
Chain: E: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
|
|
| 83 | c3i12A_ |
|
not modelled |
25.7 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: the crystal structure of the d-alanyl-alanine synthetase a from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
|
|
|
| 84 | c3u5iG_ |
|
not modelled |
25.6 |
13 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l8-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 60s subunit, ribosome b
|
|
|
| 85 | c3u9sI_ |
|
not modelled |
25.3 |
16 |
PDB header:ligase
Chain: I: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
|
|
| 86 | c3a11D_ |
|
not modelled |
25.3 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta subunit;
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
|
|
| 87 | d1e4ea2 |
|
not modelled |
25.2 |
13 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
|
|
| 88 | c3vpbC_ |
|
not modelled |
25.1 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:putative acetylornithine deacetylase;
PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
|
|
|
| 89 | c4xz7A_ |
|
not modelled |
25.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a tgase
|
|
|
| 90 | c3m3hA_ |
|
not modelled |
24.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
|
|
|
| 91 | c5b04G_ |
|
not modelled |
24.5 |
15 |
PDB header:translation
Chain: G: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 92 | d1gsoa3 |
|
not modelled |
24.3 |
27 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
|
|
| 93 | c3se7A_ |
|
not modelled |
23.8 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:vana;
PDBTitle: ancient vana
|
|
|
| 94 | c1w96B_ |
|
not modelled |
23.7 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
|
|
|
| 95 | c3j3bG_ |
|
not modelled |
23.1 |
13 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l7a;
PDBTitle: structure of the human 60s ribosomal proteins
|
|
|
| 96 | c2nv2U_ |
|
not modelled |
22.7 |
15 |
PDB header:lyase/transferase
Chain: U: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
|
|
|
| 97 | c4yakD_ |
|
not modelled |
22.2 |
13 |
PDB header:ligase
Chain: D: PDB Molecule:beta subunit of acyl-coa synthetase (ndp forming);
PDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 in complex with coenzyme a, acetyl-coenzyme a and with3 phosphorylated phosphohistidine segment (site i orientation)
|
|
|
| 98 | c5dboA_ |
|
not modelled |
21.4 |
20 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
|
|
|
| 99 | c5ax7A_ |
|
not modelled |
21.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvyl transferase 1;
PDBTitle: yeast pyruvyltransferase pvg1p
|
|
|
| 100 | d1pswa_ |
|
not modelled |
21.1 |
33 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:ADP-heptose LPS heptosyltransferase II |
|
|
| 101 | c6hxgE_ |
|
not modelled |
20.7 |
13 |
PDB header:plant protein
Chain: E: PDB Molecule:pyridoxal 5'-phosphate synthase-like subunit pdx1.2;
PDBTitle: pdx1.2/pdx1.3 complex (intermediate)
|
|
|
| 102 | c4adsF_ |
|
not modelled |
20.6 |
13 |
PDB header:transferase/transferase
Chain: F: PDB Molecule:pyridoxine biosynthetic enzyme pdx1 homologue, putative;
PDBTitle: crystal structure of plasmodial plp synthase complex
|
|
|
| 103 | c5xvsA_ |
|
not modelled |
20.5 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdp/udp-n,n'-diacetylbacillosamine 2-epimerase
PDBTitle: crystal structure of udp-glcnac 2-epimerase neuc complexed with udp
|
|
|
| 104 | d2vv5a1 |
|
not modelled |
20.3 |
23 |
Fold:Sm-like fold
Superfamily:Sm-like ribonucleoproteins
Family:Mechanosensitive channel protein MscS (YggB), middle domain |
|
|
| 105 | c1ulzA_ |
|
not modelled |
20.3 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
|
|