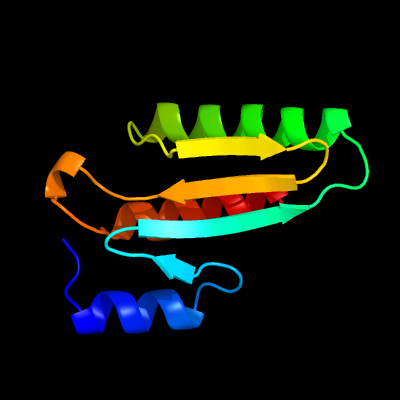
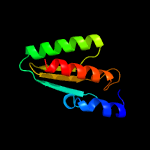
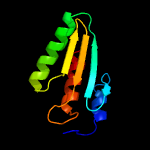
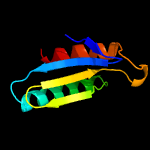
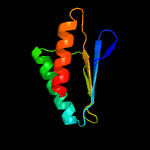
1 c2ebbA_
100.0
32
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
2 c3jstA_
100.0
36
PDB header: lyaseChain: A: PDB Molecule: putative pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of transcriptional coactivator/pterin dehydratase2 from brucella melitensis
3 d1ru0a_
100.0
29
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like
4 d1dcpa_
100.0
30
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like
5 c2v6uB_
100.0
28
PDB header: lyaseChain: B: PDB Molecule: pterin-4a-carbinolamine dehydratase;PDBTitle: high resolution crystal structure of pterin-4a-2 carbinolamine dehydratase from toxoplasma gondii
6 d1usma_
99.9
37
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like
7 c4lowA_
99.9
24
PDB header: unknown functionChain: A: PDB Molecule: acraf;PDBTitle: structure and identification of a pterin dehydratase-like protein as a2 rubisco assembly factor in the alpha-carboxysome
8 c1z9bA_
61.2
36
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: solution structure of the c1-subdomain of bacillus2 stearothermophilus translation initiation factor if2
9 c5mz2I_
59.4
24
PDB header: photosynthesisChain: I: PDB Molecule: rubisco small subunit;PDBTitle: rubisco from thalassiosira antarctica
10 d1bwvs_
59.3
24
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
11 d1a8ya3
58.0
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Calsequestrin
12 d1svdm1
55.0
18
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
13 d1bxni_
52.9
43
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
14 c5nv3P_
51.1
41
PDB header: lyaseChain: P: PDB Molecule: ribulose bisphosphate carboxylase small chain 1;PDBTitle: structure of rubisco from rhodobacter sphaeroides in complex with cabp
15 d1rbli_
49.6
23
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
16 c2ybvN_
48.6
18
PDB header: lyaseChain: N: PDB Molecule: ribulose bisphosphate carboxylase small subunit;PDBTitle: structure of rubisco from thermosynechococcus elongatus
17 d1gk8i_
48.2
41
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
18 d1ir1s_
46.7
32
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
19 d1uzhc1
43.6
41
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
20 d2v6ai1
42.3
41
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
21 d1wdds_
not modelled
42.1
27
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
22 d8ruci_
not modelled
41.7
32
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
23 d1uzdc1
not modelled
37.4
41
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
24 d1ej7s_
not modelled
36.7
36
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
25 c1qysA_
not modelled
24.3
19
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
26 c3ns6B_
not modelled
22.5
3
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: crystal structure of hte rna recognition motif of yeast eif3b residues2 76-170
27 c2jvfA_
not modelled
22.5
12
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
28 c3j4jA_
not modelled
20.9
27
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: model of full-length t. thermophilus translation initiation factor 22 refined against its cryo-em density from a 30s initiation complex map
29 c4n3gA_
not modelled
20.5
9
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5b-like protein,PDBTitle: crystal structure of eukaryotic translation initiation factor eif5b2 (870-1116) from chaetomium thermophilum, domains iii and iv
30 d1g7sa3
not modelled
19.0
19
Fold: Initiation factor IF2/eIF5b, domain 3Superfamily: Initiation factor IF2/eIF5b, domain 3Family: Initiation factor IF2/eIF5b, domain 3
31 d1w4ta1
not modelled
18.9
50
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase
32 c2kieA_
not modelled
15.7
15
PDB header: hydrolaseChain: A: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: a ph domain within ocrl bridges clathrin mediated membrane2 trafficking to phosphoinositide metabolis
33 d1mv8a1
not modelled
13.8
13
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: UDP-glucose/GDP-mannose dehydrogenase dimerisation domain
34 c4yk1A_
not modelled
13.4
20
PDB header: protein bindingChain: A: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the bid domain of bep6 from bartonella rochalimae
35 d1prtc2
not modelled
12.9
20
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Aerolysin/Pertussis toxin (APT) domain
36 c2yfwC_
not modelled
12.3
33
PDB header: cell cycleChain: C: PDB Molecule: histone h3-like centromeric protein cse4;PDBTitle: heterotetramer structure of kluyveromyces lactis cse4,h4
37 d1dlja1
not modelled
12.2
13
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: UDP-glucose/GDP-mannose dehydrogenase dimerisation domain
38 c3s6eB_
not modelled
12.1
22
PDB header: rna binding proteinChain: B: PDB Molecule: rna-binding protein 39;PDBTitle: crystal structure of a rna binding motif protein 39 (rbm39) from mus2 musculus at 0.95 a resolution
39 c4gicB_
not modelled
12.0
12
PDB header: oxidoreductaseChain: B: PDB Molecule: histidinol dehydrogenase;PDBTitle: crystal structure of a putative histidinol dehydrogenase (target psi-2 014034) from methylococcus capsulatus
40 c2hueB_
not modelled
11.6
42
PDB header: dna binding proteinChain: B: PDB Molecule: histone h3;PDBTitle: structure of the h3-h4 chaperone asf1 bound to histones h3 and h4
41 c5zojE_
not modelled
10.8
19
PDB header: dna binding proteinChain: E: PDB Molecule: inner nuclear membrane protein man1;PDBTitle: crystal structure of human smad2-man1 complex
42 c4ky3A_
not modelled
10.6
18
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or327;PDBTitle: three-dimensional structure of the orthorhombic crystal of2 computationally designed insertion domain , northeast structural3 genomics consortium (nesg) target or327
43 c2ahqA_
not modelled
10.6
14
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: solution structure of the c-terminal rpon domain of sigma-2 54 from aquifex aeolicus
44 d2j0sd1
not modelled
10.3
28
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
45 d1sqwa2
not modelled
9.6
25
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: Nip7p homolog, N-terminal domain
46 c4d10H_
not modelled
9.6
25
PDB header: signaling proteinChain: H: PDB Molecule: cop9 signalosome complex subunit 8;PDBTitle: crystal structure of the cop9 signalosome
47 c6f0fB_
not modelled
9.5
31
PDB header: chaperoneChain: B: PDB Molecule: ip2_s;PDBTitle: crystal structure asf1-ip2_s
48 c3wbkB_
not modelled
9.4
13
PDB header: biosynthetic proteinChain: B: PDB Molecule: eukaryotic translation initiation factor 5b;PDBTitle: crystal structure analysis of eukaryotic translation initiation factor2 5b and 1a complex
49 c2mztA_
not modelled
9.3
22
PDB header: rna binding proteinChain: A: PDB Molecule: protein hrb1;PDBTitle: nmr structure of the rrm3 domain of hrb1
50 d1prtb2
not modelled
8.9
20
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Aerolysin/Pertussis toxin (APT) domain
51 c2jd3B_
not modelled
8.7
26
PDB header: dna binding proteinChain: B: PDB Molecule: stbb protein;PDBTitle: parr from plasmid pb171
52 c2fhoA_
not modelled
8.6
35
PDB header: rna binding proteinChain: A: PDB Molecule: spliceosomal protein sf3b155;PDBTitle: nmr solution structure of the human spliceosomal protein2 complex p14-sf3b155
53 d1rk8a_
not modelled
8.5
31
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
54 d1tzyc_
not modelled
8.1
42
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
55 c3izyP_
not modelled
8.1
33
PDB header: rna, ribosomal proteinChain: P: PDB Molecule: translation initiation factor if-2, mitochondrial;PDBTitle: mammalian mitochondrial translation initiation factor 2
56 c5mmjv_
not modelled
8.1
31
PDB header: ribosomeChain: V: PDB Molecule: PDBTitle: structure of the small subunit of the chloroplast ribosome
57 c2qt7B_
not modelled
8.0
18
PDB header: hydrolaseChain: B: PDB Molecule: receptor-type tyrosine-protein phosphatase-like n;PDBTitle: crystallographic structure of the mature ectodomain of the human2 receptor-type protein-tyrosine phosphatase ia-2 at 1.30 angstroms
58 c5l87A_
not modelled
7.7
21
PDB header: membrane proteinChain: A: PDB Molecule: peroxin 14;PDBTitle: targeting the pex14-pex5 interaction by small molecules provides novel2 therapeutic routes to treat trypanosomiases.
59 c4kp1A_
not modelled
7.7
13
PDB header: isomeraseChain: A: PDB Molecule: isopropylmalate/citramalate isomerase large subunit;PDBTitle: crystal structure of ipm isomerase large subunit from methanococcus2 jannaschii (mj0499)
60 d1eqzg_
not modelled
7.5
42
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
61 d1o0pa_
not modelled
7.5
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
62 c3nquA_
not modelled
7.5
25
PDB header: dna binding proteinChain: A: PDB Molecule: histone h3-like centromeric protein a;PDBTitle: crystal structure of partially trypsinized (cenp-a/h4)2 heterotetramer
63 c5aonB_
not modelled
7.4
25
PDB header: signaling proteinChain: B: PDB Molecule: peroxin 14;PDBTitle: crystal structure of the conserved n-terminal domain of2 pex14 from trypanosoma brucei
64 c4kp2A_
not modelled
7.3
13
PDB header: lyaseChain: A: PDB Molecule: homoaconitase large subunit;PDBTitle: crystal structure of homoaconitase large subunit from methanococcus2 jannaschii (mj1003)
65 d1id3a_
not modelled
7.2
25
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
66 c2e44A_
not modelled
7.2
31
PDB header: rna binding proteinChain: A: PDB Molecule: insulin-like growth factor 2 mrna bindingPDBTitle: solution structure of rna binding domain in insulin-like2 growth factor 2 mrna binding protein 3
67 c3km3B_
not modelled
7.1
20
PDB header: hydrolaseChain: B: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: crystal structure of eoxycytidine triphosphate deaminase from2 anaplasma phagocytophilum at 2.1a resolution
68 c2o8kA_
not modelled
7.1
14
PDB header: transcription/dnaChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: nmr structure of the sigma-54 rpon domain bound to the-242 promoter element
69 c3nqjA_
not modelled
7.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: histone h3-like centromeric protein a;PDBTitle: crystal structure of (cenp-a/h4)2 heterotetramer
70 c2dnqA_
not modelled
6.7
12
PDB header: rna binding proteinChain: A: PDB Molecule: rna-binding protein 4b;PDBTitle: solution structure of rna binding domain 1 in rna-binding2 protein 30
71 d1vpka3
not modelled
6.7
14
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase III, beta subunit
72 d1p3ie_
not modelled
6.6
42
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
73 c2f9jP_
not modelled
6.5
38
PDB header: rna binding proteinChain: P: PDB Molecule: splicing factor 3b subunit 1;PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
74 d1rz4a1
not modelled
6.5
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Eukaryotic translation initiation factor 3 subunit 12, eIF3k, C-terminal domain
75 c3btpB_
not modelled
6.4
38
PDB header: dna binding protein, chaperoneChain: B: PDB Molecule: protein vire1;PDBTitle: crystal structure of agrobacterium tumefaciens vire2 in complex with2 its chaperone vire1: a novel fold and implications for dna binding
76 c4uqtA_
not modelled
6.4
24
PDB header: translationChain: A: PDB Molecule: u2 snrnp component ist3;PDBTitle: rrm-peptide structure in res complex
77 d2dita1
not modelled
6.4
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
78 d1whza_
not modelled
6.3
25
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: YcfA-like
79 c4v1ao_
not modelled
6.3
38
PDB header: ribosomeChain: O: PDB Molecule: PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
80 c4kjzD_
not modelled
6.2
46
PDB header: translationChain: D: PDB Molecule: translation initiation factor if-2;PDBTitle: crystal structure of thermus thermophilus if2, apo and gdp-bound forms2 (2-474)
81 c6e0cA_
not modelled
6.1
25
PDB header: nuclear proteinChain: A: PDB Molecule: histone h3-like centromeric protein a;PDBTitle: cryo-em structure of the cenp-a nucleosome (w601) in complex with a2 single chain antibody fragment
82 d1fjeb2
not modelled
6.1
12
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
83 d1kx5a_
not modelled
6.0
42
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
84 c2mt3A_
not modelled
6.0
21
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-54 factor;PDBTitle: structure of -24 dna binding domain of sigma 54 from e.coli
85 c5vbdA_
not modelled
6.0
14
PDB header: signaling proteinChain: A: PDB Molecule: usp9x;PDBTitle: crystal structure of a putative ubl domain of usp9x
86 c6an0A_
not modelled
5.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: histidinol dehydrogenase;PDBTitle: crystal structure of histidinol dehydrogenase from elizabethkingia2 anophelis
87 c4a8xB_
not modelled
5.9
33
PDB header: transcriptionChain: B: PDB Molecule: hook-like, isoform a;PDBTitle: structure of the core asap complex
88 c2jyaA_
not modelled
5.7
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1810;PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
89 c2osrA_
not modelled
5.7
27
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: nmr structure of rrm-2 of yeast npl3 protein
90 c5vldC_
not modelled
5.6
17
PDB header: oxidoreductaseChain: C: PDB Molecule: histidinol dehydrogenase, chloroplastic;PDBTitle: crystal structure of medicago truncatula l-histidinol dehydrogenase in2 complex with l-histidine and nad+
91 c2w85A_
not modelled
5.5
50
PDB header: protein transportChain: A: PDB Molecule: peroxisomal membrane anchor protein pex14;PDBTitle: structure of pex14 in complex with pex19
92 c6b4jC_
not modelled
5.4
25
PDB header: transport proteinChain: C: PDB Molecule: nucleoporin like 2;PDBTitle: crystal structure of human gle1 ctd-nup42 gbm-ddx19b(amppnp) complex
93 c2o3dA_
not modelled
5.4
11
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor, arginine/serine-rich 1;PDBTitle: structure of human sf2/asf rna recognition motif 2 (rrm2)
94 c4bptC_
not modelled
5.3
38
PDB header: oxidoreductaseChain: C: PDB Molecule: phenylalanine-4-hydroxylase (pah) (phe-4-monooxygenase);PDBTitle: structural and thermodynamic insight into phenylalanine2 hydroxylase from the human pathogen legionella pneumophila
95 d1oiaa_
not modelled
5.3
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
96 c3tcyA_
not modelled
5.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: phenylalanine-4-hydroxylase;PDBTitle: crystallographic structure of phenylalanine hydroxylase from2 chromobacterium violaceum (cpah) bound to phenylalanine in a site3 distal to the active site
97 c2jvoA_
not modelled
5.3
19
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: segmental isotope labeling of npl3
98 c6b4fD_
not modelled
5.3
25
PDB header: transport proteinChain: D: PDB Molecule: nucleoporin like 2;PDBTitle: crystal structure of human gle1 ctd-nup42 gbm complex
99 d1ivta_
not modelled
5.2
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Lamin A/C globular tail domainFamily: Lamin A/C globular tail domain