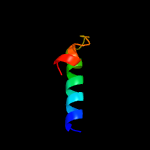| 1 |
|
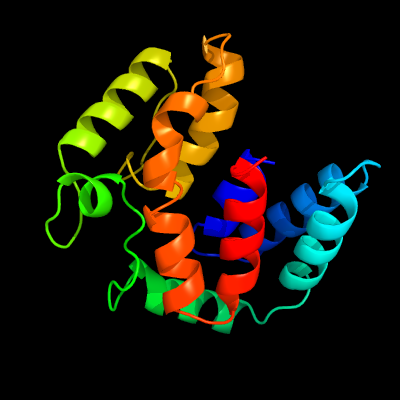
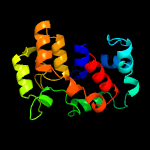
PDB 2o9x chain A domain 1
Region: 1 - 162
Aligned: 146
Modelled: 162
Confidence: 100.0%
Identity: 23%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 2 |
|
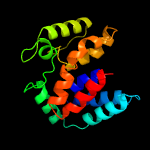
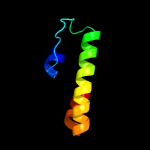
PDB 2o9x chain A
Region: 2 - 162
Aligned: 144
Modelled: 161
Confidence: 100.0%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:reductase, assembly protein;
PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
Phyre2
| 3 |
|
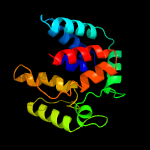
PDB 1n1c chain A
Region: 1 - 176
Aligned: 172
Modelled: 176
Confidence: 99.9%
Identity: 16%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 4 |
|
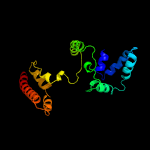
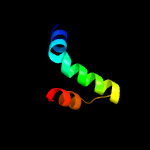
PDB 1s9u chain A
Region: 1 - 160
Aligned: 156
Modelled: 160
Confidence: 99.9%
Identity: 17%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 5 |
|
PDB 2idg chain A domain 1
Region: 89 - 141
Aligned: 52
Modelled: 52
Confidence: 98.7%
Identity: 15%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 6 |
|
PDB 5xco chain B
Region: 63 - 73
Aligned: 11
Modelled: 11
Confidence: 22.0%
Identity: 45%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:ace-arg-arg-arg-arg-cys-pro-leu-tyr-ile-ser-tyr-asp-pro-
PDBTitle: crystal structure of human k-ras g12d mutant in complex with gdp and2 cyclic inhibitory peptide
Phyre2
| 7 |
|
PDB 1lva chain A domain 3
Region: 79 - 109
Aligned: 31
Modelled: 31
Confidence: 19.7%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: C-terminal fragment of elongation factor SelB
Phyre2
| 8 |
|
PDB 2di4 chain B
Region: 122 - 159
Aligned: 38
Modelled: 38
Confidence: 16.8%
Identity: 11%
PDB header:hydrolase
Chain: B: PDB Molecule:cell division protein ftsh homolog;
PDBTitle: crystal structure of the ftsh protease domain
Phyre2
| 9 |
|
PDB 2n6v chain A
Region: 180 - 188
Aligned: 9
Modelled: 9
Confidence: 14.7%
Identity: 44%
PDB header:unknown function
Chain: A: PDB Molecule:astexin3;
PDBTitle: solution study of astexin3
Phyre2
| 10 |
|
PDB 2m8f chain A
Region: 180 - 188
Aligned: 9
Modelled: 9
Confidence: 14.5%
Identity: 44%
PDB header:unknown function
Chain: A: PDB Molecule:astexin3;
PDBTitle: structure of lasso peptide astexin3
Phyre2
| 11 |
|
PDB 2ce7 chain A domain 1
Region: 76 - 157
Aligned: 82
Modelled: 82
Confidence: 7.9%
Identity: 12%
Fold: FtsH protease domain-like
Superfamily: FtsH protease domain-like
Family: FtsH protease domain-like
Phyre2
| 12 |
|
PDB 5hbz chain E
Region: 60 - 120
Aligned: 53
Modelled: 61
Confidence: 6.3%
Identity: 21%
PDB header:hydrolase
Chain: E: PDB Molecule:non-structural protein 11;
PDBTitle: structure of eav nsp11 k170a mutant at 3.10a
Phyre2
| 13 |
|
PDB 6eb0 chain A
Region: 10 - 102
Aligned: 93
Modelled: 93
Confidence: 5.8%
Identity: 8%
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit;
PDBTitle: structure of 4-hydroxyphenylacetate 3-monooxygenase (hpab), oxygenase2 component from escherichia coli
Phyre2
| 14 |
|
PDB 3gy9 chain A
Region: 90 - 104
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:gcn5-related n-acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_001815201.1) from2 exiguobacterium sp. 255-15 at 1.52 a resolution
Phyre2
| 15 |
|
PDB 2g3a chain A domain 1
Region: 90 - 108
Aligned: 18
Modelled: 19
Confidence: 5.5%
Identity: 11%
Fold: Acyl-CoA N-acyltransferases (Nat)
Superfamily: Acyl-CoA N-acyltransferases (Nat)
Family: N-acetyl transferase, NAT
Phyre2