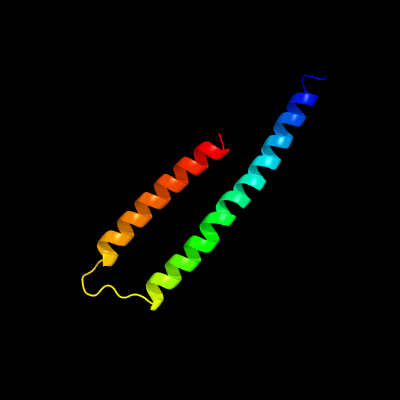
1 c4clvB_
53.5
15
PDB header: metal binding proteinChain: B: PDB Molecule: nickel-cobalt-cadmium resistance protein nccx;PDBTitle: crystal structure of dodecylphosphocholine-solubilized nccx2 from cupriavidus metallidurans 31a
2 c2n2aA_
50.5
24
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
3 d1g7da_
44.0
23
Fold: ERP29 C domain-likeSuperfamily: ERP29 C domain-likeFamily: ERP29 C domain-like
4 c1vw4a_
38.4
23
PDB header: ribosomeChain: A: PDB Molecule: PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
5 c5k57A_
27.6
12
PDB header: hydrolaseChain: A: PDB Molecule: protein ddi1 homolog 2;PDBTitle: hdd domain from human ddi2
6 c5xnlX_
24.9
46
PDB header: membrane proteinChain: X: PDB Molecule: photosystem ii reaction center protein x;PDBTitle: structure of stacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
7 d2c0ga1
24.2
19
Fold: ERP29 C domain-likeSuperfamily: ERP29 C domain-likeFamily: ERP29 C domain-like
8 c3jcux_
20.2
39
PDB header: membrane proteinChain: X: PDB Molecule: photosystem ii reaction center x protein;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
9 c3jcuX_
20.2
39
PDB header: membrane proteinChain: X: PDB Molecule: photosystem ii reaction center x protein;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
10 d1kbhb_
17.0
33
Fold: Nuclear receptor coactivator interlocking domainSuperfamily: Nuclear receptor coactivator interlocking domainFamily: Nuclear receptor coactivator interlocking domain
11 c1zhcA_
14.9
13
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein hp1242;PDBTitle: solution structure of hp1242 from helicobacter pylori
12 c3mayE_
14.0
14
PDB header: heme-binding proteinChain: E: PDB Molecule: possible exported protein;PDBTitle: crystal structure of a secreted mycobacterium tuberculosis heme-2 binding protein
13 c4cvoA_
13.8
18
PDB header: hydrolaseChain: A: PDB Molecule: dna excision repair protein ercc-6;PDBTitle: crystal structure of the n-terminal colied-coil domain of human dna2 excision repair protein ercc-6
14 c5frgA_
13.7
20
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
15 c2mnsA_
12.4
78
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p15;PDBTitle: solution nmr structure of the reovirus p15 fusion-associated small2 transmembrane (fast) protein fusion-inducing lipid packing sensor3 (flips) motif in dodecyl phosphocholine (dpc) micelles
16 c1tqeY_
10.0
38
PDB header: transcription/protein binding/dnaChain: Y: PDB Molecule: histone deacetylase 9;PDBTitle: mechanism of recruitment of class ii histone deacetylases by myocyte2 enhancer factor-2
17 c1tqeX_
10.0
38
PDB header: transcription/protein binding/dnaChain: X: PDB Molecule: histone deacetylase 9;PDBTitle: mechanism of recruitment of class ii histone deacetylases by myocyte2 enhancer factor-2
18 c2kaxA_
10.0
11
PDB header: metal binding proteinChain: A: PDB Molecule: protein s100-a5;PDBTitle: solution structure and dynamics of s100a5 in the apo and2 ca2+ -bound states
19 c2l9vA_
9.7
18
PDB header: rna binding proteinChain: A: PDB Molecule: pre-mrna-processing factor 40 homolog a;PDBTitle: nmr structure of the ff domain l24a mutant's folding transition state
20 d1a4pa_
9.4
22
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
21 c5nr5A_
not modelled
9.2
53
PDB header: dna binding proteinChain: A: PDB Molecule: mata protein;PDBTitle: nmr structure and 1h, 13c and 15n signal assignments for dictyostelium2 discoideum mata protein
22 d1e8aa_
not modelled
8.8
17
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
23 c5j1hB_
not modelled
8.2
26
PDB header: structural proteinChain: B: PDB Molecule: plectin,plectin;PDBTitle: structure of the spectrin repeats 5 and 6 of the plakin domain of2 plectin
24 c5furK_
not modelled
7.6
20
PDB header: transcriptionChain: K: PDB Molecule: transcription initiation factor tfiid subunit 6;PDBTitle: structure of human tfiid-iia bound to core promoter dna
25 c2llvA_
not modelled
7.4
20
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein sti1;PDBTitle: solution structure of the yeast sti1 dp1 domain
26 d1zfsa1
not modelled
7.1
28
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
27 d1wd3a1
not modelled
6.9
52
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Alpha-L-arabinofuranosidase B, N-terminal domain
28 c5nr6A_
not modelled
6.9
40
PDB header: dna binding proteinChain: A: PDB Molecule: matb protein;PDBTitle: nmr structure and 1h, 13c and 15n signal assignments for dictyostelium2 discoidans matb protein s71a mutant
29 c2yeqA_
not modelled
6.8
67
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase d;PDBTitle: structure of phod
30 d1hyoa1
not modelled
6.6
24
Fold: SH3-like barrelSuperfamily: Fumarylacetoacetate hydrolase, FAH, N-terminal domainFamily: Fumarylacetoacetate hydrolase, FAH, N-terminal domain
31 c2f49C_
not modelled
6.5
41
PDB header: transferaseChain: C: PDB Molecule: ste5 peptide;PDBTitle: crystal structure of fus3 in complex with a ste5 peptide
32 c2ke4A_
not modelled
6.2
18
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
33 c3eabK_
not modelled
5.9
60
PDB header: cell cycleChain: K: PDB Molecule: chmp1b;PDBTitle: crystal structure of spastin mit in complex with escrt iii
34 d1ksoa_
not modelled
5.8
17
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
35 c4uetA_
not modelled
5.7
28
PDB header: retinol-binding proteinChain: A: PDB Molecule: nematode fatty acid retinoid binding protein;PDBTitle: diversity in the structures and ligand binding sites among2 the fatty acid and retinol binding proteins of nematodes3 revealed by na-far-1 from necator americanus
36 c2y5iF_
not modelled
5.6
22
PDB header: metal-binding proteinChain: F: PDB Molecule: s100 calcium binding protein z;PDBTitle: s100z from zebrafish in complex with calcium
37 c6iczX_
not modelled
5.4
44
PDB header: splicingChain: X: PDB Molecule: prkr-interacting protein 1;PDBTitle: cryo-em structure of a human post-catalytic spliceosome (p complex) at2 3.0 angstrom
38 c2d44A_
not modelled
5.4
52
PDB header: hydrolaseChain: A: PDB Molecule: alpha-l-arabinofuranosidase b;PDBTitle: crystal structure of arabinofuranosidase complexed with2 arabinofuranosyl-alpha-1,2-xylobiose
39 c1odpA_
not modelled
5.4
36
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.6, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
40 c1odqA_
not modelled
5.4
36
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 3.7, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
41 c1odrA_
not modelled
5.4
36
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:dpc mole ratio3 of 1:40