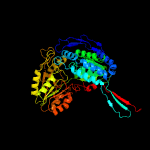
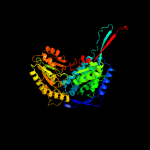
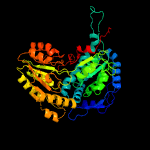
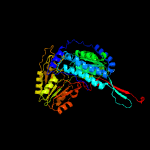
1 c4idmA_
100.0
98
PDB header: oxidoreductaseChain: A: PDB Molecule: delta-1-pyrroline-5-carboxylate dehydrogenase;PDBTitle: crystal structure of the delta-pyrroline-5-carboxylate dehydrogenase2 from mycobacterium tuberculosis
2 c4f9iA_
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase/delta-1-pyrroline-5-carboxylatePDBTitle: crystal structure of proline utilization a (puta) from geobacter2 sulfurreducens pca
3 c5ur2C_
100.0
29
PDB header: oxidoreductaseChain: C: PDB Molecule: bifunctional protein puta;PDBTitle: crystal structure of proline utilization a (puta) from bdellovibrio2 bacteriovorus inactivated by n-propargylglycine
4 c3qanB_
100.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-pyrroline-5-carboxylate dehydrogenase 1;PDBTitle: crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from2 bacillus halodurans
5 d1uzba_
100.0
29
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
6 c5kf6B_
100.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of proline utilization a from sinorhizobium meliloti2 complexed with l-tetrahydrofuroic acid and nad+ in space group p21
7 c6fk3B_
100.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure and function of aldehyde dehydrogenase from thermus2 thermophilus: an enzyme with an evolutionarily-distinct c-terminal3 arm (recombinant full-length protein in complex with propanal)
8 c3ed6B_
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
9 c6mvtA_
100.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of a bacterial aldh16 complexed with nadh
10 c3hazA_
100.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
11 c2o2qA_
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: formyltetrahydrofolate dehydrogenase;PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp
12 c3u4jB_
100.0
25
PDB header: oxidoreductaseChain: B: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 sinorhizobium meliloti
13 d1bxsa_
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
14 c4o5hD_
100.0
28
PDB header: oxidoreductaseChain: D: PDB Molecule: phenylacetaldehyde dehydrogenase;PDBTitle: x-ray crystal structure of a putative phenylacetaldehyde dehydrogenase2 from burkholderia cenocepacia
15 c2jg7G_
100.0
23
PDB header: oxidoreductaseChain: G: PDB Molecule: antiquitin;PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity
16 c2ve5H_
100.0
24
PDB header: oxidoreductaseChain: H: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystallographic structure of betaine aldehyde2 dehydrogenase from pseudomonas aeruginosa
17 d1a4sa_
100.0
22
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
18 c3iwkB_
100.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: aminoaldehyde dehydrogenase;PDBTitle: crystal structure of aminoaldehyde dehydrogenase 1 from pisum sativum2 (psamadh1)
19 c2d4eB_
100.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: 5-carboxymethyl-2-hydroxymuconate semialdehydePDBTitle: crystal structure of the hpcc from thermus thermophilus hb8
20 c4zz7E_
100.0
24
PDB header: oxidoreductaseChain: E: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of methylmalonate-semialdehyde dehydrogenase (dddc)2 from oceanimonas doudoroffii
21 d1o9ja_
not modelled
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
22 c3rh9A_
not modelled
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase (nad(p)(+));PDBTitle: the crystal structure of oxidoreductase from marinobacter aquaeolei
23 c4dalB_
not modelled
100.0
27
PDB header: oxidoreductaseChain: B: PDB Molecule: putative aldehyde dehydrogenase;PDBTitle: crystal structure of putative aldehyde dehydrogenase from2 sinorhizobium meliloti 1021
24 c5izdE_
not modelled
100.0
26
PDB header: oxidoreductaseChain: E: PDB Molecule: d-glyceraldehyde dehydrogenase (nadp(+));PDBTitle: wild-type glyceraldehyde dehydrogenase from thermoplasma acidophilum2 in complex with nadp
25 c4pt3C_
not modelled
100.0
26
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: nadph complex structure of aldehyde dehydrogenase from bacillus cereus
26 c1t90B_
not modelled
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: probable methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of methylmalonate semialdehyde dehydrogenase from2 bacillus subtilis
27 c4pxlB_
not modelled
100.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: cytosolic aldehyde dehydrogenase rf2c;PDBTitle: structure of zm aldh2-3 (rf2c) in complex with nad
28 d1wnda_
not modelled
100.0
26
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
29 c5x5uB_
not modelled
100.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: alpha-ketoglutaric semialdehyde dehydrogenase;PDBTitle: crystal strcuture of alpha-ketoglutarate-semialdehyde dehydrogenase2 (kgsadh) complexed with nad
30 c3v9iD_
not modelled
100.0
43
PDB header: oxidoreductaseChain: D: PDB Molecule: delta-1-pyrroline-5-carboxylate dehydrogenase,PDBTitle: crystal structure of human 1-pyrroline-5-carboxylate dehydrogenase2 mutant s352l
31 d1o04a_
not modelled
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
32 d1ky8a_
not modelled
100.0
25
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
33 d1ag8a_
not modelled
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
34 c4go4E_
not modelled
100.0
26
PDB header: oxidoreductaseChain: E: PDB Molecule: putative gamma-hydroxymuconic semialdehyde dehydrogenase;PDBTitle: crystal structure of pnpe in complex with nicotinamide adenine2 dinucleotide
35 c4h73E_
not modelled
100.0
23
PDB header: oxidoreductaseChain: E: PDB Molecule: aldehyde dehydrogenase;PDBTitle: thermostable aldehyde dehydrogenase from pyrobaculum sp. complexed2 with nadp+
36 c4e4gF_
not modelled
100.0
25
PDB header: oxidoreductaseChain: F: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of putative methylmalonate-semialdehyde2 dehydrogenase from sinorhizobium meliloti 1021
37 d1euha_
not modelled
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
38 c3ek1C_
not modelled
100.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from brucella2 melitensis biovar abortus 2308
39 c2w8qA_
not modelled
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase,PDBTitle: the crystal structure of human ssadh in complex with ssa.
40 c4i25B_
not modelled
100.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-aminomuconate 6-semialdehyde dehydrogenase;PDBTitle: 2.00 angstroms x-ray crystal structure of nad- and substrate-bound 2-2 aminomuconate 6-semialdehyde dehydrogenase from pseudomonas3 fluorescens
41 c4jz6A_
not modelled
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: salicylaldehyde dehydrogenase nahf;PDBTitle: crystal structure of a salicylaldehyde dehydrogenase from pseudomonas2 putida g7 complexed with salicylaldehyde
42 c3r31A_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystal structure of betaine aldehyde dehydrogenase from agrobacterium2 tumefaciens
43 c4oe4A_
not modelled
100.0
45
PDB header: oxidoreductaseChain: A: PDB Molecule: delta-1-pyrroline-5-carboxylate dehydrogenase,PDBTitle: crystal structure of yeast aldh4a1 complexed with nad+
44 c3i44A_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from bartonella2 henselae at 2.0a resolution
45 c3k2wD_
not modelled
100.0
24
PDB header: oxidoreductaseChain: D: PDB Molecule: betaine-aldehyde dehydrogenase;PDBTitle: crystal structure of betaine-aldehyde dehydrogenase from2 pseudoalteromonas atlantica t6c
46 c3ifgH_
not modelled
100.0
24
PDB header: oxidoreductaseChain: H: PDB Molecule: succinate-semialdehyde dehydrogenase (nadp+);PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2
47 c5vbfH_
not modelled
100.0
24
PDB header: oxidoreductaseChain: H: PDB Molecule: nad-dependent succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of succinate semialdehyde dehydrogenase from2 burkholderia vietnamiensis
48 c4knaA_
not modelled
100.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: n-succinylglutamate 5-semialdehyde dehydrogenase;PDBTitle: crystal structure of an n-succinylglutamate 5-semialdehyde2 dehydrogenase from burkholderia thailandensis
49 c3b4wA_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis aldehyde dehydrogenase2 complexed with nad+
50 c5u0mB_
not modelled
100.0
25
PDB header: oxidoreductaseChain: B: PDB Molecule: n-succinylglutamate 5-semialdehyde dehydrogenase;PDBTitle: fatty aldehyde dehydrogenase from marinobacter aquaeolei vt8 and2 cofactor complex
51 c4pxnB_
not modelled
100.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: structure of zm aldh7 in complex with nad
52 c4qyjD_
not modelled
100.0
27
PDB header: oxidoreductaseChain: D: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of phenylacetaldehyde dehydrogenase from pseudomonas putida2 s12
53 c4yweE_
not modelled
100.0
26
PDB header: oxidoreductaseChain: E: PDB Molecule: putative aldehyde dehydrogenase;PDBTitle: crystal structure of a putative aldehyde dehydrogenase from2 burkholderia cenocepacia
54 c4i3wC_
not modelled
100.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase (nad+);PDBTitle: structure of phosphonoacetaldehyde dehydrogenase in complex with2 gylceraldehyde-3-phosphate and cofactor nad+
55 c3jz4C_
not modelled
100.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate-semialdehyde dehydrogenase [nadp+];PDBTitle: crystal structure of e. coli nadp dependent enzyme
56 c2hg2A_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase a;PDBTitle: structure of lactaldehyde dehydrogenase
57 c3prlD_
not modelled
100.0
25
PDB header: oxidoreductaseChain: D: PDB Molecule: nadp-dependent glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-phosphate2 dehydrogenase from bacillus halodurans c-125
58 c6dbbA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: putative aldehyde dehydrogenase family protein;PDBTitle: crystal structure of a putative aldehyde dehydrogenase family protein2 burkholderia cenocepacia j2315 in complex with partially reduced nadh
59 c5j6bB_
not modelled
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from burkholderia2 thailandensis in covelent complex with nadph
60 c5mz5A_
not modelled
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: aldh21);PDBTitle: crystal structure of aldehyde dehydrogenase 21 (aldh21) from2 physcomitrella patens in its apoform
61 c2vroB_
not modelled
100.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from2 burkholderia xenovorans lb400
62 c4lihG_
not modelled
100.0
25
PDB header: oxidoreductaseChain: G: PDB Molecule: gamma-glutamyl-gamma-aminobutyraldehyde dehydrogenase;PDBTitle: the crystal structure of gamma-glutamyl-gamma-aminobutyraldehyde2 dehydrogenase from burkholderia cenocepacia j2315
63 c3ju8B_
not modelled
100.0
27
PDB header: oxidoreductaseChain: B: PDB Molecule: succinylglutamic semialdehyde dehydrogenase;PDBTitle: crystal structure of succinylglutamic semialdehyde dehydrogenase from2 pseudomonas aeruginosa.
64 c5ux5C_
not modelled
100.0
26
PDB header: oxidoreductase/transferaseChain: C: PDB Molecule: bifunctional protein proline utilization a (puta);PDBTitle: structure of proline utilization a (puta) from corynebacterium2 freiburgense
65 d1bi9a_
not modelled
100.0
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
66 c4h7nA_
not modelled
100.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: the structure of putative aldehyde dehydrogenase puta from anabaena2 variabilis.
67 c4ohtB_
not modelled
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of succinic semialdehyde dehydrogenase from2 streptococcus pyogenes in complex with nadp+ as the cofactor
68 c4itaA_
not modelled
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase;PDBTitle: structure of bacterial enzyme in complex with cofactor
69 c3vz0B_
not modelled
100.0
25
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nad-dependent aldehyde dehydrogenase;PDBTitle: structural insights into cofactor and substrate selection by gox0499
70 c3rosA_
not modelled
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 lactobacillus acidophilus
71 c3efvC_
not modelled
100.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: putative succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of a putative succinate-semialdehyde dehydrogenase2 from salmonella typhimurium lt2 with bound nad
72 c3r64A_
not modelled
100.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: nad dependent benzaldehyde dehydrogenase;PDBTitle: crystal structure of a nad-dependent benzaldehyde dehydrogenase from2 corynebacterium glutamicum
73 c6d97B_
not modelled
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase 12;PDBTitle: structure of aldehyde dehydrogenase 12 (aldh12) from zea mays
74 c3pqaA_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: lactaldehyde dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase gapn2 from methanocaldococcus jannaschii dsm 2661
75 c5tjrE_
not modelled
100.0
24
PDB header: oxidoreductaseChain: E: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: x-ray crystal structure of a methylmalonate semialdehyde dehydrogenase2 from pseudomonas sp. aac
76 c5fhzF_
not modelled
100.0
26
PDB header: oxidoreductaseChain: F: PDB Molecule: aldehyde dehydrogenase family 1 member a3;PDBTitle: human aldehyde dehydrogenase 1a3 complexed with nad(+) and retinoic2 acid
77 d1ad3a_
not modelled
100.0
20
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
78 c4qgkB_
not modelled
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: fatty aldehyde dehydrogenase;PDBTitle: structure of the human sjogren larsson syndrome enzyme fatty aldehyde2 dehydrogenase (faldh)
79 c5nnoA_
not modelled
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of tbaldh3 complexed with nad and an3057 aldehyde
80 c3v4cB_
not modelled
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase (nadp+);PDBTitle: crystal structure of a semialdehyde dehydrogenase from sinorhizobium2 meliloti 1021
81 d1ez0a_
not modelled
100.0
19
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
82 c5iuuA_
not modelled
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase family protein;PDBTitle: crystal structure of indole-3-acetaldehyde dehydrogenase in apo form
83 c4dngB_
not modelled
100.0
25
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized aldehyde dehydrogenase aldy;PDBTitle: crystal structure of putative aldehyde dehydrogenase from bacillus2 subtilis subsp. subtilis str. 168
84 c5ujuA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 burkholderia multivorans
85 c3lnsD_
not modelled
100.0
21
PDB header: oxidoreductaseChain: D: PDB Molecule: benzaldehyde dehydrogenase;PDBTitle: benzaldehyde dehydrogenase, a class 3 aldehyde dehydrogenase, with2 bound nadp+ and benzoate adduct
86 c5j78B_
not modelled
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: acetaldehyde dehydrogenase (acetylating);PDBTitle: crystal structure of an acetylating aldehyde dehydrogenase from2 geobacillus thermoglucosidasius
87 c3k9dD_
not modelled
100.0
17
PDB header: oxidoreductaseChain: D: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of probable aldehyde dehydrogenase from listeria2 monocytogenes egd-e
88 c4c3sA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of a propionaldehyde dehydrogenase from the clostridium2 phytofermentans fucose utilisation bacterial microcompartment
89 d1o20a_
not modelled
100.0
13
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
90 c5jfnA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of rhodopseudomonas palustris propionaldehyde2 dehydrogenase with bound coa and acylated cys330
91 c4jbeA_
not modelled
100.0
13
PDB header: oxidoreductaseChain: A: PDB Molecule: gamma-glutamyl phosphate reductase;PDBTitle: 1.95 angstrom crystal structure of gamma-glutamyl phosphate reductase2 from saccharomonospora viridis.
92 c3my7A_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase/acetaldehyde dehydrogenase;PDBTitle: the crystal structure of the acdh domain of an alcohol dehydrogenase2 from vibrio parahaemolyticus to 2.25a
93 c4ghkB_
not modelled
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: gamma-glutamyl phosphate reductase;PDBTitle: x-ray crystal structure of gamma-glutamyl phosphate reductase from2 burkholderia thailandensis
94 d1vlua_
not modelled
100.0
15
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
95 c2h5gA_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: delta 1-pyrroline-5-carboxylate synthetase;PDBTitle: crystal structure of human pyrroline-5-carboxylate synthetase
96 c1vluB_
not modelled
100.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: gamma-glutamyl phosphate reductase;PDBTitle: crystal structure of gamma-glutamyl phosphate reductase (yor323c) from2 saccharomyces cerevisiae at 2.40 a resolution
97 d1k75a_
not modelled
96.2
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: L-histidinol dehydrogenase HisD
98 c6an0A_
not modelled
95.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: histidinol dehydrogenase;PDBTitle: crystal structure of histidinol dehydrogenase from elizabethkingia2 anophelis
99 c5vldC_
not modelled
94.7
14
PDB header: oxidoreductaseChain: C: PDB Molecule: histidinol dehydrogenase, chloroplastic;PDBTitle: crystal structure of medicago truncatula l-histidinol dehydrogenase in2 complex with l-histidine and nad+
100 c4gicB_
not modelled
92.2
16
PDB header: oxidoreductaseChain: B: PDB Molecule: histidinol dehydrogenase;PDBTitle: crystal structure of a putative histidinol dehydrogenase (target psi-2 014034) from methylococcus capsulatus
101 c4g07A_
not modelled
87.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: histidinol dehydrogenase;PDBTitle: the crystal structure of the c366s mutant of hdh from brucella suis
102 c3v4gA_
not modelled
60.4
19
PDB header: dna binding proteinChain: A: PDB Molecule: arginine repressor;PDBTitle: 1.60 angstrom resolution crystal structure of an arginine repressor2 from vibrio vulnificus cmcp6
103 d1y5ea1
not modelled
57.6
21
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
104 d1uz5a3
not modelled
46.5
7
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like
105 c2yvqA_
not modelled
41.7
8
PDB header: ligaseChain: A: PDB Molecule: carbamoyl-phosphate synthase;PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
106 d1mkza_
not modelled
38.7
16
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
107 d2g2ca1
not modelled
38.7
16
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
108 d2f7wa1
not modelled
32.3
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
109 d2ftsa3
not modelled
29.0
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like
110 c1uz5A_
not modelled
28.4
7
PDB header: molybdopterin biosynthesisChain: A: PDB Molecule: 402aa long hypothetical molybdopterinPDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
111 d1a9xa2
not modelled
28.2
23
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain
112 c6eqoB_
not modelled
28.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: acetyl-coenzyme a synthetase;PDBTitle: tri-functional propionyl-coa synthase of erythrobacter sp. nap1 with2 bound nadp+ and phosphomethylphosphonic acid adenylate ester
113 d1xxaa_
not modelled
27.1
30
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor
114 d1r7la_
not modelled
26.5
20
Fold: Bacillus phage proteinSuperfamily: Bacillus phage proteinFamily: Bacillus phage protein
115 c3cu2A_
not modelled
26.0
13
PDB header: isomeraseChain: A: PDB Molecule: ribulose-5-phosphate 3-epimerase;PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
116 c2is8A_
not modelled
25.4
18
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
117 c2yukA_
not modelled
24.5
22
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the hmg box of human myeloid/lymphoid2 or mixed-lineage leukemia protein 3 homolog
118 c5g2rA_
not modelled
21.8
10
PDB header: transferaseChain: A: PDB Molecule: molybdopterin biosynthesis protein cnx1;PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana