1 c2ekgB_
100.0
37
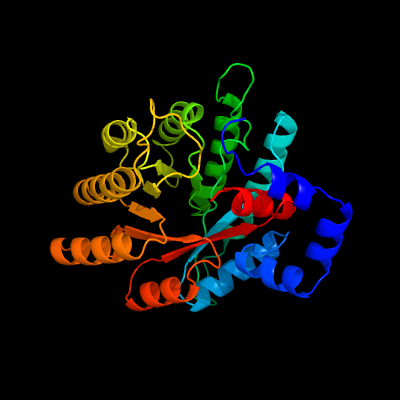
PDB header: oxidoreductaseChain: B: PDB Molecule: proline dehydrogenase/delta-1-pyrroline-5-carboxylatePDBTitle: structure of thermus thermophilus proline dehydrogenase inactivated by2 n-propargylglycine
2 c5ur2C_
100.0
25
PDB header: oxidoreductaseChain: C: PDB Molecule: bifunctional protein puta;PDBTitle: crystal structure of proline utilization a (puta) from bdellovibrio2 bacteriovorus inactivated by n-propargylglycine
3 c4f9iA_
100.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase/delta-1-pyrroline-5-carboxylatePDBTitle: crystal structure of proline utilization a (puta) from geobacter2 sulfurreducens pca
4 c4h6rA_
100.0
39
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: structure of reduced deinococcus radiodurans proline dehydrogenase
5 c1k87A_
100.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of e.coli puta (residues 1-669)
6 c3e2sA_
100.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure reduced puta86-630 mutant y540s complexed with l-2 proline
7 c1tj2A_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional puta protein;PDBTitle: crystal structure of e. coli puta proline dehydrogenase domain2 (residues 86-669) complexed with acetate
8 c3hazA_
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
9 d1tj1a2
100.0
28
Fold: TIM beta/alpha-barrelSuperfamily: FAD-linked oxidoreductaseFamily: Proline dehydrohenase domain of bifunctional PutA protein
10 c5kf6B_
100.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of proline utilization a from sinorhizobium meliloti2 complexed with l-tetrahydrofuroic acid and nad+ in space group p21
11 c5ux5C_
100.0
21
PDB header: oxidoreductase/transferaseChain: C: PDB Molecule: bifunctional protein proline utilization a (puta);PDBTitle: structure of proline utilization a (puta) from corynebacterium2 freiburgense
12 c3itgA_
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional protein puta;PDBTitle: structure the proline utilization a proline dehydrogenase2 domain (puta86-630) inactivated with n-propargylglycine
13 c5tnvA_
96.3
17
PDB header: isomeraseChain: A: PDB Molecule: ap endonuclease, family protein 2;PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
14 d1tz9a_
93.2
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: UxuA-like
15 c4k3zA_
92.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
16 c4ovxA_
91.3
18
PDB header: isomeraseChain: A: PDB Molecule: xylose isomerase domain protein tim barrel;PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
17 c3dx5A_
90.7
17
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
18 c3cnyA_
89.7
12
PDB header: biosynthetic proteinChain: A: PDB Molecule: inositol catabolism protein iole;PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
19 c5hmqE_
87.6
16
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxyphenylpyruvate dioxygenase;PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
20 d1qt1a_
86.1
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
21 c2hk1D_
not modelled
85.8
13
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
22 d1yx1a1
not modelled
84.5
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like
23 c3vylB_
not modelled
82.6
14
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose 3-epimerase;PDBTitle: structure of l-ribulose 3-epimerase
24 c2qw5B_
not modelled
81.5
14
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
25 c3ktcB_
not modelled
77.3
17
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
26 d1i60a_
not modelled
77.1
16
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
27 c5zfsA_
not modelled
76.2
20
PDB header: isomeraseChain: A: PDB Molecule: d-allulose-3-epimerase;PDBTitle: crystal structure of arthrobacter globiformis m30 sugar epimerase2 which can produce d-allulose from d-fructose
28 c2zdsB_
not modelled
76.0
18
PDB header: dna binding proteinChain: B: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of sco6571 from streptomyces coelicolor a3(2)
29 c3ju2A_
not modelled
74.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein smc04130;PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
30 d2glka1
not modelled
73.3
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
31 c2ou4C_
not modelled
71.9
14
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
32 d1xima_
not modelled
71.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
33 c3vniC_
not modelled
68.2
13
PDB header: isomeraseChain: C: PDB Molecule: xylose isomerase domain protein tim barrel;PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
34 d1kkoa1
not modelled
66.9
17
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like
35 c3qxbB_
not modelled
66.0
12
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
36 c1kczA_
not modelled
62.7
19
PDB header: lyaseChain: A: PDB Molecule: beta-methylaspartase;PDBTitle: crystal structure of beta-methylaspartase from clostridium2 tetanomorphum. mg-complex.
37 d2q02a1
not modelled
61.6
8
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
38 d1bxba_
not modelled
60.9
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
39 c2qpuB_
not modelled
60.3
14
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amylase type a isozyme;PDBTitle: sugar tongs mutant s378p in complex with acarbose
40 c3bdkB_
not modelled
58.5
18
PDB header: lyaseChain: B: PDB Molecule: d-mannonate dehydratase;PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
41 c3wqoB_
not modelled
56.6
10
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein mj1311;PDBTitle: crystal structure of d-tagatose 3-epimerase-like protein
42 d2pb1a1
not modelled
56.4
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
43 c3icgD_
not modelled
52.2
11
PDB header: hydrolaseChain: D: PDB Molecule: endoglucanase d;PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
44 c3p6lA_
not modelled
52.0
6
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
45 d1bxca_
not modelled
51.5
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
46 c3kwsB_
not modelled
49.0
17
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
47 d1ht6a2
not modelled
48.3
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
48 d2guya2
not modelled
45.7
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
49 d1avaa2
not modelled
44.0
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
50 d1k77a_
not modelled
43.3
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Hypothetical protein YgbM (EC1530)
51 c5oydA_
not modelled
42.6
13
PDB header: hydrolaseChain: A: PDB Molecule: cellulase, putative, cel5d;PDBTitle: gh5 endo-xyloglucanase from cellvibrio japonicus
52 c4w8aA_
not modelled
41.7
11
PDB header: hydrolaseChain: A: PDB Molecule: exo-xyloglucanase;PDBTitle: crystal structure of xeg5b, a gh5 xyloglucan-specific beta-1,4-2 glucanase from ruminal metagenomic library, in the native form
53 c3l23A_
not modelled
38.3
10
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
54 d1muwa_
not modelled
33.6
17
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
55 c3l55B_
not modelled
32.0
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: b-1,4-endoglucanase/cellulase;PDBTitle: crystal structure of a putative beta-1,4-endoglucanase / cellulase2 from prevotella bryantii
56 c3cqkB_
not modelled
30.4
13
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose-5-phosphate 3-epimerase ulae;PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
57 c1uz4A_
not modelled
29.4
15
PDB header: hydrolaseChain: A: PDB Molecule: man5a;PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
58 d1uuqa_
not modelled
29.4
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
59 c6nbmB_
not modelled
29.0
22
PDB header: lyaseChain: B: PDB Molecule: enolase;PDBTitle: crystal structure of enolase from legionella pneumophila bound to2 phosphate and magnesium
60 d1mxga2
not modelled
28.2
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
61 c2oylB_
not modelled
27.8
19
PDB header: hydrolaseChain: B: PDB Molecule: endoglycoceramidase ii;PDBTitle: endo-glycoceramidase ii from rhodococcus sp.: cellobiose-like2 imidazole complex
62 c4m0xB_
not modelled
27.6
16
PDB header: isomeraseChain: B: PDB Molecule: chloromuconate cycloisomerase;PDBTitle: crystal structure of 2-chloromuconate cycloisomerase from rhodococcus2 opacus 1cp
63 c4e8gA_
not modelled
27.4
24
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, n-terminalPDBTitle: crystal structure of an enolase (mandelate racemase subgroup) from2 paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg
64 d1vjza_
not modelled
27.2
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
65 d1edga_
not modelled
26.9
8
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
66 c2zvrA_
not modelled
26.6
10
PDB header: isomeraseChain: A: PDB Molecule: uncharacterized protein tm_0416;PDBTitle: crystal structure of a d-tagatose 3-epimerase-related protein from2 thermotoga maritima
67 c2yl8A_
not modelled
26.6
16
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
68 c3zviB_
not modelled
25.8
19
PDB header: lyaseChain: B: PDB Molecule: methylaspartate ammonia-lyase;PDBTitle: methylaspartate ammonia lyase from clostridium tetanomorphum mutant2 l384a
69 d1ur4a_
not modelled
25.8
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
70 c4nf7A_
not modelled
25.7
14
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-glucanase cel5c;PDBTitle: crystal structure of the gh5 family catalytic domain of endo-1,4-beta-2 glucanase cel5c from butyrivibrio proteoclasticus.
71 c1qhoA_
not modelled
25.4
23
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
72 c4yheB_
not modelled
25.4
19
PDB header: hydrolaseChain: B: PDB Molecule: gh5;PDBTitle: native bacteroidetes-affiliated gh5 cellulase linked with a2 polysaccharide utilization locus
73 c3bmwA_
not modelled
25.0
14
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
74 c4x0vH_
not modelled
24.3
12
PDB header: hydrolaseChain: H: PDB Molecule: beta-1,3-1,4-glucanase;PDBTitle: structure of a gh5 family lichenase from caldicellulosiruptor sp. f32
75 c3sjnB_
not modelled
24.3
24
PDB header: lyaseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of enolase spea_3858 (target efi-500646) from2 shewanella pealeana with magnesium bound
76 d1hx0a2
not modelled
24.1
32
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
77 c3t8qA_
not modelled
24.1
13
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 family protein from hoeflea phototrophica
78 d1cgta4
not modelled
24.1
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
79 c5ccuA_
not modelled
23.9
23
PDB header: hydrolaseChain: A: PDB Molecule: putative secreted endoglycosylceramidase;PDBTitle: crystal structure of endoglycoceramidase i from rhodococ-cus equi
80 c1cygA_
not modelled
23.8
23
PDB header: glycosyltransferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
81 c3aysA_
not modelled
23.7
17
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: gh5 endoglucanase from a ruminal fungus in complex with cellotriose
82 c2zunB_
not modelled
23.3
20
PDB header: hydrolaseChain: B: PDB Molecule: 458aa long hypothetical endo-1,4-beta-glucanase;PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii
83 c2jepB_
not modelled
23.3
10
PDB header: hydrolaseChain: B: PDB Molecule: xyloglucanase;PDBTitle: native family 5 xyloglucanase from paenibacillus pabuli
84 c3ncoA_
not modelled
23.2
20
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase fncel5a;PDBTitle: crystal structure of fncel5a from f. nodosum rt17-b1
85 c2bg5C_
not modelled
22.9
19
PDB header: transferaseChain: C: PDB Molecule: phosphoenolpyruvate-protein kinase;PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme i-domain2 from the thermoanaerobacter tengcongensis pep: sugar3 phosphotransferase system (pts)
86 d1jpma1
not modelled
22.4
12
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like
87 c2y8kA_
not modelled
22.3
10
PDB header: hydrolaseChain: A: PDB Molecule: carbohydrate binding family 6;PDBTitle: structure of ctgh5-cbm6, an arabinoxylan-specific xylanase.
88 c4ee9A_
not modelled
22.0
12
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: crystal structure of the rbcel1 endo-1,4-glucanase
89 c1kkoB_
not modelled
21.8
18
PDB header: lyaseChain: B: PDB Molecule: 3-methylaspartate ammonia-lyase;PDBTitle: crystal structure of citrobacter amalonaticus2 methylaspartate ammonia lyase
90 c5y6tA_
not modelled
21.8
10
PDB header: carbohydrateChain: A: PDB Molecule: endo-1,4-beta-mannanase;PDBTitle: crystal structure of endo-1,4-beta-mannanase from eisenia fetida
91 d1cxla4
not modelled
21.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
92 c5zxgB_
not modelled
21.4
23
PDB header: hydrolaseChain: B: PDB Molecule: cyclic maltosyl-maltose hydrolase;PDBTitle: cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from arthrobacter2 globiformis, ligand-free form
93 d1gcya2
not modelled
21.4
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
94 c5dlcC_
not modelled
21.0
19
PDB header: transferaseChain: C: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
95 d2ffca1
not modelled
20.3
6
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
96 d1ob0a2
not modelled
20.2
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
97 d2g0wa1
not modelled
20.0
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
98 d1pama4
not modelled
19.7
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
99 d1wu7a1
not modelled
19.2
13
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS