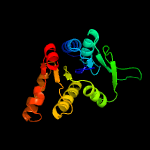
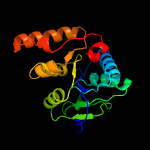
1 d1ydha_
100.0
27
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
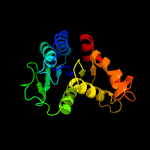
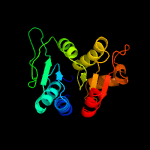
2 c5zbjA_
100.0
42
PDB header: hydrolaseChain: A: PDB Molecule: putative cytokinin riboside 5'-monophosphatePDBTitle: crystal strcuture of type-i log from pseudomonas aeruginosa pao1
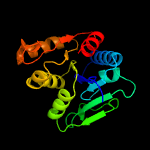
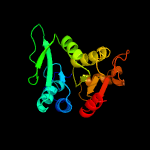
3 c5itsD_
100.0
33
PDB header: hydrolaseChain: D: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of log from corynebacterium glutamicum
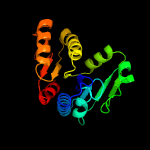
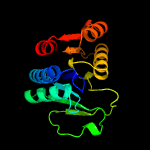
4 d1t35a_
100.0
28
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
5 c2q4dB_
100.0
28
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lysine decarboxylase-like protein at5g11950;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
6 c3quaA_
100.0
73
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein and possible2 molybdenum cofactor protein from mycobacterium smegmatis
7 c3sbxC_
100.0
83
PDB header: unknown functionChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium marinum bound to adenosine 5'-monophosphate amp
8 c5ajtA_
100.0
32
PDB header: hydrolaseChain: A: PDB Molecule: phosphoribohydrolase lonely guy;PDBTitle: crystal structure of ligand-free phosphoribohydrolase lonely guy from2 claviceps purpurea
9 d2q4oa1
100.0
30
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
10 c2q4oA_
100.0
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein at2g37210/t2n18.3;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
11 c5zi9B_
100.0
26
PDB header: hydrolaseChain: B: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of type-ii log from streptomyces coelicolor a3
12 d1weka_
100.0
26
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
13 c5wq3A_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of type-ii log from corynebacterium glutamicum
14 d1weha_
100.0
29
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
15 c3bq9A_
100.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted rossmann fold nucleotide-binding domain-PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
16 c6gfmA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: pyrimidine/purine nucleotide 5'-monophosphate nucleosidase;PDBTitle: crystal structure of the escherichia coli nucleosidase ppnn (pppgpp-2 form)
17 c3gh1A_
100.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted nucleotide-binding protein;PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
18 c1rcuB_
100.0
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein vt76;PDBTitle: x-ray structure of tm1055 northeast structural genomics2 consortium target vt76
19 d1rcua_
100.0
22
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
20 c2iz6A_
100.0
28
PDB header: metal transportChain: A: PDB Molecule: molybdenum cofactor carrier protein;PDBTitle: structure of the chlamydomonas rheinhardtii moco carrier2 protein
21 c3uqzB_
not modelled
99.6
20
PDB header: dna binding proteinChain: B: PDB Molecule: dna processing protein dpra;PDBTitle: x-ray structure of dna processing protein a (dpra) from streptococcus2 pneumoniae
22 c3majA_
not modelled
99.6
17
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a;PDBTitle: crystal structure of putative dna processing protein dpra from2 rhodopseudomonas palustris cga009
23 c4ljkA_
not modelled
99.6
18
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a (dpra);PDBTitle: structural insights into the unique single-stranded dna binding mode2 of dna processing protein a from helicobacter pylori
24 d2nx2a1
not modelled
98.5
17
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: YpsA-like
25 c6d73C_
not modelled
97.4
24
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel, subfamily m;PDBTitle: cryo-em structure of the zebrafish trpm2 channel in the presence of2 ca2+
26 c6mizC_
not modelled
97.4
25
PDB header: membrane proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: human trpm2 ion channel in an adpr-bound state
27 c6d73B_
not modelled
97.2
24
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel, subfamily m;PDBTitle: cryo-em structure of the zebrafish trpm2 channel in the presence of2 ca2+
28 c6co7C_
not modelled
97.2
23
PDB header: membrane proteinChain: C: PDB Molecule: predicted protein;PDBTitle: structure of the nvtrpm2 channel in complex with ca2+
29 c6bcqB_
not modelled
96.9
24
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of trpm4 in atp bound state with long coiled coil at2 3.3 angstrom resolution
30 c6bcoD_
not modelled
96.9
24
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of trpm4 in atp bound state with short coiled coil2 at 2.9 angstrom resolution
31 c6drkD_
not modelled
96.7
25
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel, subfamily m,PDBTitle: structure of trpm2 ion channel receptor by single particle electron2 cryo-microscopy, apo state
32 c3imkA_
not modelled
96.5
16
PDB header: metal binding proteinChain: A: PDB Molecule: putative molybdenum carrier protein;PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
33 c6nr3D_
not modelled
93.4
16
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
34 c6nr3C_
not modelled
93.4
16
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
35 c6nr3B_
not modelled
93.4
16
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
36 c6nr3A_
not modelled
93.4
16
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
37 c5vtoA_
not modelled
93.4
27
PDB header: hydrolaseChain: A: PDB Molecule: blasticidin m;PDBTitle: solution structure of blsm
38 c4jenB_
not modelled
93.2
20
PDB header: hydrolaseChain: B: PDB Molecule: cmp n-glycosidase;PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
39 c6evsA_
not modelled
92.4
17
PDB header: transferaseChain: A: PDB Molecule: n-deoxyribosyltransferase;PDBTitle: characterization of 2-deoxyribosyltransferase from psychrotolerant2 bacterium bacillus psychrosaccharolyticus: a suitable biocatalyst for3 the industrial synthesis of antiviral and antitumoral nucleosides
40 c2khzB_
not modelled
92.2
19
PDB header: nuclear proteinChain: B: PDB Molecule: c-myc-responsive protein rcl;PDBTitle: solution structure of rcl
41 c6nr2B_
not modelled
91.8
19
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
42 c6nr2C_
not modelled
91.8
19
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
43 c6nr2D_
not modelled
91.8
19
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
44 c6nr2A_
not modelled
91.8
19
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
45 d2nu7b1
not modelled
91.3
18
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains
46 c5nbrB_
not modelled
88.7
18
PDB header: transferaseChain: B: PDB Molecule: deoxyribosyltransferase;PDBTitle: 2-desoxiribosyltransferase from leishmania mexicana
47 c3bgkA_
not modelled
88.0
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the crystal structure of hypothetic protein smu.573 from streptococcus2 mutans
48 c2r3bA_
not modelled
87.6
17
PDB header: transferaseChain: A: PDB Molecule: yjef-related protein;PDBTitle: crystal structure of a ribokinase-like superfamily protein (ef1790)2 from enterococcus faecalis v583 at 1.80 a resolution
49 c4jemA_
not modelled
86.5
17
PDB header: hydrolaseChain: A: PDB Molecule: cmp/hydroxymethyl cmp hydrolase;PDBTitle: crystal structure of milb complexed with cytidine 5'-monophosphate
50 c3zlbA_
not modelled
81.2
17
PDB header: transferaseChain: A: PDB Molecule: phosphoglycerate kinase;PDBTitle: crystal structure of phosphoglycerate kinase from streptococcus2 pneumoniae
51 c4qjiB_
not modelled
79.1
16
PDB header: ligaseChain: B: PDB Molecule: phosphopantothenate--cysteine ligase;PDBTitle: crystal structure of the c-terminal ctp-binding domain of a2 phosphopantothenoylcysteine decarboxylase/phosphopantothenate-3 cysteine ligase with bound ctp from mycobacterium smegmatis
52 d1f8ya_
not modelled
77.4
23
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
53 d1u0ta_
not modelled
76.3
23
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: NAD kinase-like
54 c2jzcA_
not modelled
75.6
12
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine transferase subunitPDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
55 c2bonB_
not modelled
74.6
26
PDB header: transferaseChain: B: PDB Molecule: lipid kinase;PDBTitle: structure of an escherichia coli lipid kinase (yegs)
56 c1zrsB_
not modelled
72.3
25
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: wild-type ld-carboxypeptidase
57 c1zmrA_
not modelled
72.3
18
PDB header: transferaseChain: A: PDB Molecule: phosphoglycerate kinase;PDBTitle: crystal structure of the e. coli phosphoglycerate kinase
58 c6efwA_
not modelled
72.2
19
PDB header: lyaseChain: A: PDB Molecule: atp-dependent (s)-nad(p)h-hydrate dehydratase;PDBTitle: crystal structure of a yjef family protein from cryptococcus2 neoformans var. grubii serotype a
59 d2auna2
not modelled
72.1
29
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like
60 d1phpa_
not modelled
67.3
18
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
61 c3q3vA_
not modelled
66.8
15
PDB header: transferaseChain: A: PDB Molecule: phosphoglycerate kinase;PDBTitle: crystal structure of phosphoglycerate kinase from campylobacter2 jejuni.
62 d1yoba1
not modelled
65.4
24
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
63 c3ehdA_
not modelled
63.6
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of conserved protein from enterococcus faecalis v583
64 c4ng4B_
not modelled
60.0
16
PDB header: transferaseChain: B: PDB Molecule: phosphoglycerate kinase;PDBTitle: structure of phosphoglycerate kinase (cbu_1782) from coxiella burnetii
65 c3s40C_
not modelled
57.2
24
PDB header: transferaseChain: C: PDB Molecule: diacylglycerol kinase;PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
66 c2qv7A_
not modelled
57.0
17
PDB header: transferaseChain: A: PDB Molecule: diacylglycerol kinase dgkb;PDBTitle: crystal structure of diacylglycerol kinase dgkb in complex with adp2 and mg
67 c4as2D_
not modelled
55.9
25
PDB header: hydrolaseChain: D: PDB Molecule: phosphorylcholine phosphatase;PDBTitle: pseudomonas aeruginosa phosphorylcholine phosphatase. monoclinic form
68 d2jgra1
not modelled
54.1
26
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
69 d2bona1
not modelled
53.7
26
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
70 d1v6sa_
not modelled
52.4
13
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
71 c3d3jA_
not modelled
51.2
17
PDB header: protein bindingChain: A: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of human edc3p
72 d1s2da_
not modelled
51.2
10
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
73 c4e5sC_
not modelled
51.1
14
PDB header: hydrolaseChain: C: PDB Molecule: mccflike protein (ba_5613);PDBTitle: crystal structure of mccflike protein (ba_5613) from bacillus2 anthracis str. ames
74 d1e4ea1
not modelled
50.5
22
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain
75 d1fw8a_
not modelled
50.1
19
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
76 d2f62a1
not modelled
49.7
20
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
77 c4dg5A_
not modelled
48.7
15
PDB header: transferaseChain: A: PDB Molecule: phosphoglycerate kinase;PDBTitle: crystal structure of staphylococcal phosphoglycerate kinase
78 c4h1hB_
not modelled
47.3
15
PDB header: hydrolaseChain: B: PDB Molecule: lmo1638 protein;PDBTitle: crystal structure of mccf homolog from listeria monocytogenes egd-e
79 d1iowa1
not modelled
45.1
22
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain
80 c6fsiA_
not modelled
45.0
12
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: crystal structure of semiquinone flavodoxin 1 from bacillus cereus2 (1.32 a resolution)
81 c6bpqD_
not modelled
44.8
11
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
82 c6bpqB_
not modelled
44.8
11
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
83 c6bpqC_
not modelled
44.8
11
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
84 c6bpqA_
not modelled
44.8
11
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
85 c3d3kD_
not modelled
44.8
18
PDB header: protein bindingChain: D: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of human edc3p
86 c5f2kA_
not modelled
44.7
31
PDB header: transferaseChain: A: PDB Molecule: fatty acid o-methyltransferase;PDBTitle: crystal structure of mycobacterial fatty acid o-methyltransferase in2 complex with sah and octanoate
87 c2wc1A_
not modelled
43.5
19
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
88 c3c5yD_
not modelled
43.0
13
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
89 d1qpga_
not modelled
42.6
19
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
90 d2p1ra1
not modelled
42.3
21
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
91 c2dg2D_
not modelled
42.2
17
PDB header: protein bindingChain: D: PDB Molecule: apolipoprotein a-i binding protein;PDBTitle: crystal structure of mouse apolipoprotein a-i binding protein
92 d1hdia_
not modelled
41.2
15
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
93 d1ehia1
not modelled
41.1
23
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain
94 c2i80B_
not modelled
40.8
19
PDB header: ligaseChain: B: PDB Molecule: d-alanine-d-alanine ligase;PDBTitle: allosteric inhibition of staphylococcus aureus d-alanine:d-alanine2 ligase revealed by crystallographic studies
95 c5ohxB_
not modelled
40.7
17
PDB header: lyaseChain: B: PDB Molecule: cystathionine beta-synthase;PDBTitle: structure of active cystathionine b-synthase from apis mellifera
96 d2qv7a1
not modelled
39.5
15
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
97 c2duwA_
not modelled
39.4
13
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
98 c5b3kA_
not modelled
39.3
19
PDB header: electron transportChain: A: PDB Molecule: uncharacterized protein pa3435;PDBTitle: c101a mutant of flavodoxin from pseudomonas aeruginosa
99 c3gndC_
not modelled
38.8
15
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
100 d1u7za_
not modelled
38.6
16
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like
101 c4ei8A_
not modelled
38.3
8
PDB header: replicationChain: A: PDB Molecule: plasmid replication protein repx;PDBTitle: crystal structure of bacillus cereus tubz, apo-form
102 c4nguA_
not modelled
38.3
6
PDB header: transport proteinChain: A: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 desulfovibrio alaskensis g20 (dde_1548), target efi-510103, with3 bound d-ala-d-ala
103 c3hy5A_
not modelled
37.6
12
PDB header: transport proteinChain: A: PDB Molecule: retinaldehyde-binding protein 1;PDBTitle: crystal structure of cralbp
104 c5dmxC_
not modelled
37.0
24
PDB header: ligaseChain: C: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase from acinetobacter2 baumannii, space group p212121
105 d1ag9a_
not modelled
36.9
27
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
106 c3k3pA_
not modelled
36.2
19
PDB header: ligaseChain: A: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of the apo form of d-alanine:d-alanine ligase (ddl)2 from streptococcus mutans
107 c4fu0B_
not modelled
35.8
19
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase 7;PDBTitle: crystal structure of vang d-ala:d-ser ligase from enterococcus2 faecalis
108 c4imrA_
not modelled
34.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-oxoacyl-(acyl-carrier-protein) reductase;PDBTitle: crystal structure of 3-oxoacyl (acyl-carrier-protein) reductase2 (target efi-506442) from agrobacterium tumefaciens c58 with nadp3 bound
109 c6odmC_
not modelled
34.6
36
PDB header: viral proteinChain: C: PDB Molecule: capsid vertex component 1;PDBTitle: herpes simplex virus type 1 (hsv-1) portal vertex-adjacent2 capsid/catc, asymmetric unit
110 c4heqB_
not modelled
34.4
9
PDB header: electron transportChain: B: PDB Molecule: flavodoxin;PDBTitle: the crystal structure of flavodoxin from desulfovibrio gigas
111 d1m6ex_
not modelled
34.2
12
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Salicylic acid carboxyl methyltransferase (SAMT)
112 d2fcra_
not modelled
34.1
19
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
113 d1ja1a2
not modelled
33.9
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like
114 c3imfA_
not modelled
33.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain dehydrogenase;PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
115 c3tlgB_
not modelled
33.6
23
PDB header: hydrolaseChain: B: PDB Molecule: mccf;PDBTitle: microcin c7 self immunity protein mccf in the inactive mutant apo2 state
116 c6c8rA_
not modelled
33.4
15
PDB header: transferaseChain: A: PDB Molecule: loganic acid o-methyltransferase;PDBTitle: loganic acid o-methyltransferase complexed with sah and loganic acid
117 c3qjgD_
not modelled
33.4
17
PDB header: oxidoreductaseChain: D: PDB Molecule: epidermin biosynthesis protein epid;PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
118 d16pka_
not modelled
32.9
16
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase
119 d1vmea1
not modelled
32.8
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
120 c3nywD_
not modelled
32.8
16
PDB header: oxidoreductaseChain: D: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a betaketoacyl-[acp] reductase (fabg) from2 bacteroides thetaiotaomicron