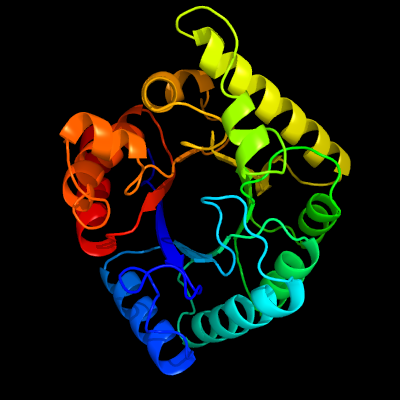
| 1 | c2bmbA_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase from3 saccharomyces cerevisiae
|
|
|
|
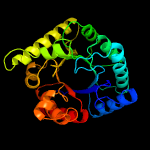
| 2 | d1ajza_
|
|
 |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
|
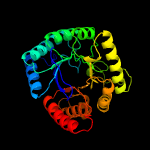
| 3 | c5z79F_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: F: PDB Molecule:hydroxymethyldihydropterin pyrophosphokinase-
PDBTitle: crystal structure analysis of the hppk-dhps in complex with substrates
|
|
|
|
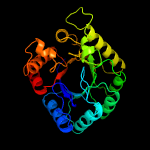
| 4 | c6omzA_
|
|
 |
100.0 |
37 |
PDB header:ligase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from mycobacterium2 smegmatis with bound 6-hydroxymethylpterin-monophosphate
|
|
|
|
| 5 | c5uswD_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: D: PDB Molecule:dihydropteroate synthase;
PDBTitle: the crystal structure of 7,8-dihydropteroate synthase from vibrio2 fischeri es114
|
|
|
|
| 6 | c3tr9A_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
|
|
|
| 7 | c5visB_
|
|
 |
100.0 |
31 |
PDB header:hydrolase,oxidoreductase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: 1.73 angstrom resolution crystal structure of dihydropteroate synthase2 (folp-smz_b27) from soil uncultured bacterium.
|
|
|
|
| 8 | d1ad1a_
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
|
| 9 | d1eyea_
|
|
 |
100.0 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
|
| 10 | c1tx2A_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
|
|
|
| 11 | d1tx2a_
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
|
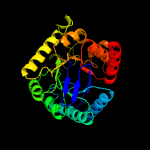
| 12 | c5uurA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: xanthomonas albilineans dihydropteroate synthase with 4-aminobenzoic2 acid
|
|
|
|
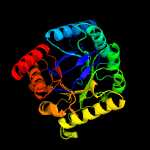
| 13 | c2y5sA_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
|
|
|
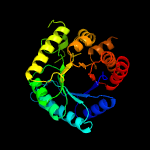
| 14 | c2vefB_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
|
|
|
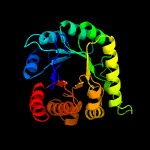
| 15 | c2dzaA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
|
|
|
| 16 | c2vp8A_
|
|
 |
100.0 |
98 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
|
|
|
| 17 | c6cluC_
|
|
 |
100.0 |
37 |
PDB header:antimicrobial protein
Chain: C: PDB Molecule:dihydropteroate synthase;
PDBTitle: staphylococcus aureus dihydropteroate synthase (sadhps) f17l e208k2 double mutant structure
|
|
|
|
| 18 | c3mcnA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
|
|
|
| 19 | c2yciX_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
|
|
|
| 20 | d1f6ya_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
|
|
|
| 21 | d3bofa1 |
|
not modelled |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
|
|
| 22 | c2h9aB_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-sulfur protein;
PDBTitle: corrinoid iron-sulfur protein
|
|
|
| 23 | c4o1fB_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase dhps;
PDBTitle: structure of a methyltransferase component in complex with thf2 involved in o-demethylation
|
|
|
| 24 | c3bolB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
|
|
| 25 | c4cczA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: crystal structure of human 5-methyltetrahydrofolate-homocysteine2 methyltransferase, the homocysteine and folate binding domains
|
|
|
| 26 | c3k13A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
|
|
| 27 | c5vooB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate homocysteine s-methyltransferase;
PDBTitle: methionine synthase folate-binding domain with methyltetrahydrofolate2 from thermus thermophilus hb8
|
|
|
| 28 | c4djdD_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase/vitamin-binding protein
Chain: D: PDB Molecule:corrinoid/iron-sulfur protein small subunit;
PDBTitle: crystal structure of folate-free corrinoid iron-sulfur protein (cfesp)2 in complex with its methyltransferase (metr)
|
|
|
| 29 | c4mwaA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
| 30 | c4djeE_ |
|
not modelled |
99.8 |
19 |
PDB header:transferase/vitamin-binding protein
Chain: E: PDB Molecule:corrinoid/iron-sulfur protein large subunit;
PDBTitle: crystal structure of folate-bound corrinoid iron-sulfur protein2 (cfesp) in complex with its methyltransferase (metr), co-crystallized3 with folate
|
|
|
| 31 | c2h9aA_ |
|
not modelled |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-sulfur
PDBTitle: corrinoid iron-sulfur protein
|
|
|
| 32 | c2yclA_ |
|
not modelled |
99.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-sulfur
PDBTitle: complete structure of the corrinoid,iron-sulfur protein including2 the n-terminal domain with a 4fe-4s cluster
|
|
|
| 33 | c3noyA_ |
|
not modelled |
99.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
|
|
| 34 | c2y0fD_ |
|
not modelled |
98.5 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
|
|
| 35 | c3ivuB_ |
|
not modelled |
97.8 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
|
|
| 36 | c3t4cD_ |
|
not modelled |
97.8 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 1;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
|
|
|
| 37 | d1qopa_ |
|
not modelled |
97.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 38 | c3bg3B_ |
|
not modelled |
97.7 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
| 39 | d1nvma2 |
|
not modelled |
97.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 40 | d1vr6a1 |
|
not modelled |
97.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
|
|
| 41 | c4jn6C_ |
|
not modelled |
97.5 |
18 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
|
|
|
| 42 | c1zfjA_ |
|
not modelled |
97.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
|
|
| 43 | c3bg3A_ |
|
not modelled |
97.5 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
| 44 | c1nvmG_ |
|
not modelled |
97.5 |
15 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
| 45 | c3fs2A_ |
|
not modelled |
97.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 bruciella melitensis at 1.85a resolution
|
|
|
| 46 | c5kzmA_ |
|
not modelled |
97.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 47 | d1wbha1 |
|
not modelled |
97.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 48 | c4e38A_ |
|
not modelled |
97.3 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 49 | d1mxsa_ |
|
not modelled |
97.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 50 | c5n2pA_ |
|
not modelled |
97.2 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 51 | c4lu0A_ |
|
not modelled |
97.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-keto-3-deoxy-d-manno-octulosonate-8-phosphate2 synthase from pseudomonas aeruginosa.
|
|
|
| 52 | c6mdyC_ |
|
not modelled |
97.2 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a 2-dehydro-3-deoxyphosphooctonate aldolase from2 legionella pneumophila philadelphia 1
|
|
|
| 53 | c1vs1B_ |
|
not modelled |
97.2 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
|
|
| 54 | c6oviA_ |
|
not modelled |
97.2 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 55 | c5ey5A_ |
|
not modelled |
97.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 56 | c3navB_ |
|
not modelled |
97.0 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 57 | c3thaB_ |
|
not modelled |
97.0 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
| 58 | c4lrtC_ |
|
not modelled |
97.0 |
21 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
|
|
|
| 59 | d1vhca_ |
|
not modelled |
97.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 60 | c1ps9A_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-dienoyl2 coa reductase
|
|
|
| 61 | c4wcxC_ |
|
not modelled |
97.0 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
| 62 | c3nvtA_ |
|
not modelled |
97.0 |
20 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
|
|
| 63 | d1ps9a1 |
|
not modelled |
96.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 64 | c2ftpA_ |
|
not modelled |
96.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
|
|
| 65 | c1rr2A_ |
|
not modelled |
96.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
|
|
| 66 | c2ekcA_ |
|
not modelled |
96.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 67 | c3pg8B_ |
|
not modelled |
96.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
|
|
| 68 | c3vndD_ |
|
not modelled |
96.9 |
21 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 69 | c4qccA_ |
|
not modelled |
96.8 |
19 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 70 | c3sz8D_ |
|
not modelled |
96.8 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
|
|
| 71 | c1zcoA_ |
|
not modelled |
96.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
|
|
| 72 | d1geqa_ |
|
not modelled |
96.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 73 | d1xcfa_ |
|
not modelled |
96.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 74 | c3ajxA_ |
|
not modelled |
96.7 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:3-hexulose-6-phosphate synthase;
PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
|
|
|
| 75 | d1q6oa_ |
|
not modelled |
96.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 76 | c4z87B_ |
|
not modelled |
96.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: structure of the imp dehydrogenase from ashbya gossypii bound to gdp
|
|
|
| 77 | c1ydoC_ |
|
not modelled |
96.5 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
|
|
| 78 | c3bleA_ |
|
not modelled |
96.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in complexed with2 malonate
|
|
|
| 79 | c6qkgB_ |
|
not modelled |
96.5 |
18 |
PDB header:flavoprotein
Chain: B: PDB Molecule:ncr a;
PDBTitle: 2-naphthoyl-coa reductase(ncr)
|
|
|
| 80 | c3labA_ |
|
not modelled |
96.4 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase;
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
|
|
| 81 | c3stgA_ |
|
not modelled |
96.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
|
|
| 82 | c2infB_ |
|
not modelled |
96.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from bacillus2 subtilis
|
|
|
| 83 | c4bk9B_ |
|
not modelled |
96.3 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxo
PDBTitle: crystal structure of 2-keto-3-deoxy-6-phospho-gluconate aldolase from2 zymomonas mobilis atcc 29191
|
|
|
| 84 | c3js3C_ |
|
not modelled |
96.2 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
|
|
| 85 | c3gr7A_ |
|
not modelled |
96.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 86 | d1rd5a_ |
|
not modelled |
96.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 87 | c3w9zA_ |
|
not modelled |
96.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 88 | c3hf3A_ |
|
not modelled |
96.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
|
|
| 89 | c3cixA_ |
|
not modelled |
96.2 |
14 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
|
|
| 90 | c4qslC_ |
|
not modelled |
96.1 |
18 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
|
|
|
| 91 | c2h90A_ |
|
not modelled |
96.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
|
|
| 92 | d1vhna_ |
|
not modelled |
96.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 93 | c4qslE_ |
|
not modelled |
96.0 |
19 |
PDB header:ligase
Chain: E: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
|
|
|
| 94 | c3jr2D_ |
|
not modelled |
96.0 |
26 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:hexulose-6-phosphate synthase sgbh;
PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 95 | c4ph6A_ |
|
not modelled |
96.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: structure of 3-dehydroquinate dehydratase from enterococcus faecalis
|
|
|
| 96 | c4rtbA_ |
|
not modelled |
96.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
| 97 | c5k9xA_ |
|
not modelled |
95.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
| 98 | c4ff0B_ |
|
not modelled |
95.9 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion2 mutant, complexed with imp
|
|
|
| 99 | c3bg5C_ |
|
not modelled |
95.9 |
18 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
|
|
|
| 100 | c4k3zA_ |
|
not modelled |
95.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-erythrulose 4-phosphate dehydrogenase;
PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
|
|
|
| 101 | c4nu7C_ |
|
not modelled |
95.9 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 102 | c5tchG_ |
|
not modelled |
95.9 |
22 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 103 | d1rqba2 |
|
not modelled |
95.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 104 | c4fxsA_ |
|
not modelled |
95.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 105 | c5ocsB_ |
|
not modelled |
95.7 |
20 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 106 | c3d0cB_ |
|
not modelled |
95.7 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 107 | c3e96B_ |
|
not modelled |
95.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 108 | c4q33F_ |
|
not modelled |
95.7 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine 5'-monophosphate dehydrogenase from2 clostridium perfringens complexed with imp and a110
|
|
|
| 109 | c2cw6B_ |
|
not modelled |
95.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
|
|
| 110 | c3a9iA_ |
|
not modelled |
95.6 |
20 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with lys
|
|
|
| 111 | c4zr8B_ |
|
not modelled |
95.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: structure of uroporphyrinogen decarboxylase from acinetobacter2 baumannii
|
|
|
| 112 | d1losc_ |
|
not modelled |
95.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 113 | d1z41a1 |
|
not modelled |
95.5 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 114 | c4xp7A_ |
|
not modelled |
95.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine(20) synthase [nad(p)+]-like;
PDBTitle: crystal structure of human trna dihydrouridine synthase 2
|
|
|
| 115 | d1d9ea_ |
|
not modelled |
95.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
|
|
| 116 | c6ei9A_ |
|
not modelled |
95.5 |
14 |
PDB header:flavoprotein
Chain: A: PDB Molecule:trna-dihydrouridine synthase b;
PDBTitle: crystal structure of e. coli trna-dihydrouridine synthase b (dusb)
|
|
|
| 117 | c3rmjB_ |
|
not modelled |
95.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
|
|
|
| 118 | c3tsdA_ |
|
not modelled |
95.5 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase from2 bacillus anthracis str. ames complexed with xmp
|
|
|
| 119 | c3ru6C_ |
|
not modelled |
95.4 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of orotidine 5'-phosphate2 decarboxylase (pyrf) from campylobacter jejuni subsp. jejuni nctc3 11168
|
|
|
| 120 | c3b0vD_ |
|
not modelled |
95.4 |
19 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
|
|