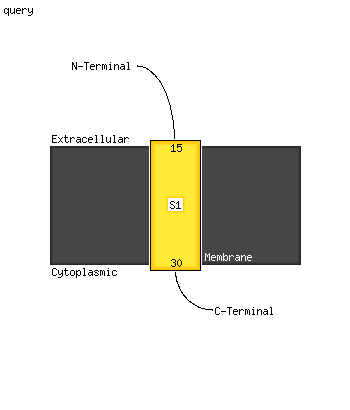
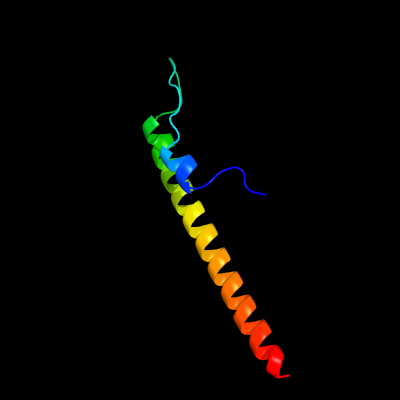
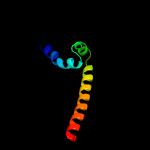
1 c4ug1A_
99.7
21
PDB header: cell cycleChain: A: PDB Molecule: cell cycle protein gpsb;PDBTitle: gpsb n-terminal domain
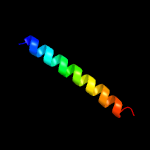
2 c6gqaD_
99.6
23
PDB header: cell cycleChain: D: PDB Molecule: cell cycle protein gpsb;PDBTitle: cell division regulator s. pneumoniae gpsb
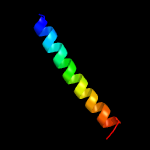
3 c2wukD_
99.6
22
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
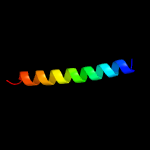
4 c4ug3C_
99.6
18
PDB header: cell cycleChain: C: PDB Molecule: cell cycle protein gpsb;PDBTitle: b. subtilis gpsb n-terminal domain
5 c5fv8A_
89.5
24
PDB header: structural proteinChain: A: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
6 c5fv8B_
86.6
24
PDB header: structural proteinChain: B: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
7 c4l9uA_
76.5
14
PDB header: signaling proteinChain: A: PDB Molecule: ras guanyl-releasing protein 1;PDBTitle: structure of c-terminal coiled coil of rasgrp1
8 c3gp4B_
76.4
16
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal structure of putative merr family transcriptional regulator2 from listeria monocytogenes
9 c2ke4A_
75.9
12
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
10 c3iynQ_
75.9
24
PDB header: virusChain: Q: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
11 c2vz4A_
75.1
25
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional activator tipa;PDBTitle: the n-terminal domain of merr-like protein tipal bound to promoter dna
12 c1cosC_
66.4
28
PDB header: alpha-helical bundleChain: C: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
13 c3gpvA_
65.8
7
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal structure of a transcriptional regulator, merr2 family from bacillus thuringiensis
14 c5wlqA_
65.7
17
PDB header: motor proteinChain: A: PDB Molecule: capsid assembly scaffolding protein,myosin-7,microtubule-PDBTitle: crystal structure of amino acids 1677-1755 of human beta cardiac2 myosin fused to gp7 and eb1
15 c1cosA_
65.5
28
PDB header: alpha-helical bundleChain: A: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
16 c1cosB_
64.2
28
PDB header: alpha-helical bundleChain: B: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
17 c3iynR_
63.7
24
PDB header: virusChain: R: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
18 c3vkhD_
63.3
15
PDB header: motor proteinChain: D: PDB Molecule: PDBTitle: x-ray structure of a functional full-length dynein motor domain
19 c2x6pA_
61.0
28
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
20 c2x6pB_
60.9
28
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
21 c3qo8A_
not modelled
60.4
20
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase, cytoplasmic;PDBTitle: crystal structure of seryl-trna synthetase from candida albicans
22 c2x6pC_
not modelled
59.6
28
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
23 c1debA_
not modelled
59.4
19
PDB header: structural proteinChain: A: PDB Molecule: adenomatous polyposis coli protein;PDBTitle: crystal structure of the n-terminal coiled coil domain from apc
24 c4l9uB_
not modelled
57.7
16
PDB header: signaling proteinChain: B: PDB Molecule: ras guanyl-releasing protein 1;PDBTitle: structure of c-terminal coiled coil of rasgrp1
25 c3cvfA_
not modelled
57.3
9
PDB header: signaling proteinChain: A: PDB Molecule: homer protein homolog 3;PDBTitle: crystal structure of the carboxy terminus of homer3
26 d1q08a_
not modelled
56.4
18
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators
27 c2mxrA_
not modelled
56.2
15
PDB header: signaling proteinChain: A: PDB Molecule: protein phosphatase 1 regulatory subunit 12a;PDBTitle: solution structure of coiled coil domain of myosin binding subunit of2 myosin light chain phosphatase
28 c1t6fA_
not modelled
55.4
15
PDB header: cell cycleChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of the coiled-coil dimerization motif of2 geminin
29 c2jgoA_
not modelled
54.9
24
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
30 c3ljmC_
not modelled
54.9
24
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
31 c3ljmA_
not modelled
54.9
24
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
32 c2jgoB_
not modelled
54.9
24
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
33 d1q06a_
not modelled
54.3
10
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators
34 c5k92A_
not modelled
53.7
28
PDB header: de novo proteinChain: A: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
35 c5k92C_
not modelled
53.7
28
PDB header: de novo proteinChain: C: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
36 c3ljmB_
not modelled
53.4
24
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
37 c2jgoC_
not modelled
53.4
24
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
38 c1coiA_
not modelled
52.8
24
PDB header: alpha-helical bundleChain: A: PDB Molecule: coil-vald;PDBTitle: designed trimeric coiled coil-vald
39 c5k92B_
not modelled
52.2
28
PDB header: de novo proteinChain: B: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
40 c6aklA_
not modelled
51.2
13
PDB header: protein bindingChain: A: PDB Molecule: suppressor of ikbke 1;PDBTitle: crystal structure of striatin3 in complex with sike1 coiled-coil2 domain
41 c2e43A_
not modelled
50.1
9
PDB header: transcription/dnaChain: A: PDB Molecule: ccaat/enhancer-binding protein beta;PDBTitle: crystal structure of c/ebpbeta bzip homodimer k269a mutant2 bound to a high affinity dna fragment
42 c1fosF_
not modelled
49.5
11
PDB header: transcription/dnaChain: F: PDB Molecule: c-jun proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
43 d1nkpa_
not modelled
47.9
11
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain
44 c3ra3D_
not modelled
47.8
15
PDB header: de novo proteinChain: D: PDB Molecule: p2f;PDBTitle: crystal structure of a section of a de novo design gigadalton protein2 fibre
45 c3n4xB_
not modelled
46.5
19
PDB header: replicationChain: B: PDB Molecule: monopolin complex subunit csm1;PDBTitle: structure of csm1 full-length
46 c1t2kD_
not modelled
46.1
17
PDB header: transcription/dnaChain: D: PDB Molecule: cyclic-amp-dependent transcription factor atf-2;PDBTitle: structure of the dna binding domains of irf3, atf-2 and jun2 bound to dna
47 c3m9bK_
not modelled
45.7
12
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
48 c3rrkA_
not modelled
45.7
19
PDB header: proton transportChain: A: PDB Molecule: v-type atpase 116 kda subunit;PDBTitle: crystal structure of the cytoplasmic n-terminal domain of subunit i,2 homolog of subunit a, of v-atpase
49 d1r05a_
not modelled
45.5
14
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain
50 c1hf9B_
not modelled
44.7
9
PDB header: atpase inhibitorChain: B: PDB Molecule: atpase inhibitor (mitochondrial);PDBTitle: c-terminal coiled-coil domain from bovine if1
51 c1jccC_
not modelled
43.5
10
PDB header: membrane proteinChain: C: PDB Molecule: major outer membrane lipoprotein;PDBTitle: crystal structure of a novel alanine-zipper trimer at 1.7 a2 resolution, v13a,l16a,v20a,l23a,v27a,m30a,v34a mutations
52 c6akkA_
not modelled
43.3
13
PDB header: protein bindingChain: A: PDB Molecule: suppressor of ikbke 1;PDBTitle: crystal structure of the second coiled-coil domain of sike1
53 c2o7hF_
not modelled
42.7
19
PDB header: transcriptionChain: F: PDB Molecule: general control protein gcn4;PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
54 c6g3eA_
not modelled
41.7
16
PDB header: lyaseChain: A: PDB Molecule: argininosuccinate lyase;PDBTitle: crystal structure of edds lyase in complex with formate
55 c3vkgA_
not modelled
40.2
15
PDB header: motor proteinChain: A: PDB Molecule: dynein heavy chain, cytoplasmic;PDBTitle: x-ray structure of an mtbd truncation mutant of dynein motor domain
56 c3ra3B_
not modelled
39.1
17
PDB header: de novo proteinChain: B: PDB Molecule: p2f;PDBTitle: crystal structure of a section of a de novo design gigadalton protein2 fibre
57 c5kb0A_
not modelled
39.0
30
PDB header: de novo proteinChain: A: PDB Molecule: pb(ii)zn(ii)(grand coil ser-l16cl30h)3+;PDBTitle: crystal structure of a tris-thiolate pb(ii) complex in a de novo2 three-stranded coiled coil peptide
58 c1aq5C_
not modelled
38.8
16
PDB header: coiled-coilChain: C: PDB Molecule: cartilage matrix protein;PDBTitle: high-resolution solution nmr structure of the trimeric coiled-coil2 domain of chicken cartilage matrix protein, 20 structures
59 c1aq5B_
not modelled
38.8
16
PDB header: coiled-coilChain: B: PDB Molecule: cartilage matrix protein;PDBTitle: high-resolution solution nmr structure of the trimeric coiled-coil2 domain of chicken cartilage matrix protein, 20 structures
60 c1ci6A_
not modelled
38.5
17
PDB header: transcriptionChain: A: PDB Molecule: transcription factor atf-4;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
61 c3kinB_
not modelled
38.5
16
PDB header: motor proteinChain: B: PDB Molecule: kinesin heavy chain;PDBTitle: kinesin (dimeric) from rattus norvegicus
62 d1jnra1
not modelled
37.0
20
Fold: Spectrin repeat-likeSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
63 c1go4F_
not modelled
36.9
19
PDB header: cell cycleChain: F: PDB Molecule: mitotic spindle assembly checkpoint protein mad1;PDBTitle: crystal structure of mad1-mad2 reveals a conserved mad2 binding motif2 in mad1 and cdc20.
64 c1g6uB_
not modelled
36.5
31
PDB header: de novo proteinChain: B: PDB Molecule: domain swapped dimer;PDBTitle: crystal structure of a domain swapped dimer
65 c3vr8E_
not modelled
36.4
16
PDB header: oxidoreductaseChain: E: PDB Molecule: flavoprotein subunit of complex ii;PDBTitle: mitochondrial rhodoquinol-fumarate reductase from the parasitic2 nematode ascaris suum
66 c2zhhA_
not modelled
36.1
15
PDB header: transcriptionChain: A: PDB Molecule: redox-sensitive transcriptional activator soxr;PDBTitle: crystal structure of soxr
67 c1ij2C_
not modelled
35.9
15
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
68 c1ij3C_
not modelled
35.5
15
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
69 c1ij3B_
not modelled
35.5
15
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
70 c1rb6C_
not modelled
35.5
15
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as n16a2 tetragonal form
71 c3k7zA_
not modelled
35.5
15
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
72 c1rb1A_
not modelled
35.5
15
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
73 c3k7zB_
not modelled
35.5
15
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
74 c1rb1B_
not modelled
35.5
15
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
75 c1swiA_
not modelled
35.5
15
PDB header: leucine zipperChain: A: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with benzene
76 c6mctL_
not modelled
35.0
50
PDB header: de novo proteinChain: L: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
77 c6mctF_
not modelled
35.0
50
PDB header: de novo proteinChain: F: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
78 c6mctI_
not modelled
35.0
50
PDB header: de novo proteinChain: I: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
79 c6mq2D_
not modelled
35.0
50
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
80 c6mctB_
not modelled
35.0
50
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
81 c6mctA_
not modelled
35.0
50
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
82 c6mctJ_
not modelled
35.0
50
PDB header: de novo proteinChain: J: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
83 c6mctG_
not modelled
35.0
50
PDB header: de novo proteinChain: G: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
84 c6mctE_
not modelled
35.0
50
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
85 c6mctN_
not modelled
35.0
50
PDB header: de novo proteinChain: N: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
86 c6mctH_
not modelled
35.0
50
PDB header: de novo proteinChain: H: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
87 c6mctM_
not modelled
35.0
50
PDB header: de novo proteinChain: M: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
88 c6mctO_
not modelled
35.0
50
PDB header: de novo proteinChain: O: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
89 c6mpwA_
not modelled
35.0
50
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
90 c6mctD_
not modelled
35.0
50
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
91 c6mctK_
not modelled
35.0
50
PDB header: de novo proteinChain: K: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
92 c6mctC_
not modelled
35.0
50
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
93 c5gpeB_
not modelled
34.6
12
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, merr-family;PDBTitle: crystal structure of the transcription regulator pbrr691 from2 ralstonia metallidurans ch34 in complex with lead(ii)
94 c6nr92_
not modelled
34.6
16
PDB header: chaperoneChain: 2: PDB Molecule: prefoldin subunit 2;PDBTitle: htric-hpfd class5
95 c6mpwC_
not modelled
34.4
50
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
96 c6mq2B_
not modelled
34.4
50
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
97 c6mpwD_
not modelled
34.4
50
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
98 c6mq2E_
not modelled
34.4
50
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
99 c6mpwB_
not modelled
34.4
50
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
100 c6mq2C_
not modelled
34.4
50
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
101 c6mpwE_
not modelled
34.4
50
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
102 c6mq2A_
not modelled
34.4
50
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
103 c2yy0D_
not modelled
33.2
21
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
104 c1fosE_
not modelled
32.9
14
PDB header: transcription/dnaChain: E: PDB Molecule: p55-c-fos proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
105 c1ci6B_
not modelled
32.9
11
PDB header: transcriptionChain: B: PDB Molecule: transcription factor c/ebp beta;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
106 c4y66C_
not modelled
32.9
23
PDB header: cell cycleChain: C: PDB Molecule: mnd1;PDBTitle: crystal structure of giardia lamblia hop2-mnd1 complex
107 c3a5tB_
not modelled
32.8
17
PDB header: transcription regulator/dnaChain: B: PDB Molecule: transcription factor mafg;PDBTitle: crystal structure of mafg-dna complex
108 c1ij2B_
not modelled
32.6
15
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
109 c5zk1A_
not modelled
32.4
10
PDB header: transcription/dnaChain: A: PDB Molecule: cyclic amp-responsive element-binding protein 1;PDBTitle: crystal structure of the crtc2(semet)-creb-cre complex
110 c2gl2B_
not modelled
32.3
16
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,y69g) of2 bacterial adhesin fada
111 c3qaoA_
not modelled
32.1
16
PDB header: transcription regulatorChain: A: PDB Molecule: merr-like transcriptional regulator;PDBTitle: the crystal structure of the n-terminal domain of a merr-like2 transcriptional regulator from listeria monocytogenes egd-e
112 c1ysaD_
not modelled
31.9
13
PDB header: transcription/dnaChain: D: PDB Molecule: protein (gcn4);PDBTitle: the gcn4 basic region leucine zipper binds dna as a dimer2 of uninterrupted alpha helices: crystal structure of the3 protein-dna complex
113 c4om3D_
not modelled
31.7
20
PDB header: transcription, dna bindingChain: D: PDB Molecule: transducin-like enhancer protein 1;PDBTitle: crystal structure of human tle1 q-domain residues 20-156
114 c4y66D_
not modelled
31.3
14
PDB header: cell cycleChain: D: PDB Molecule: putative tbpip family protein;PDBTitle: crystal structure of giardia lamblia hop2-mnd1 complex
115 d1r8da_
not modelled
31.2
16
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators
116 c2wt7B_
not modelled
30.8
20
PDB header: transcriptionChain: B: PDB Molecule: transcription factor mafb;PDBTitle: crystal structure of the bzip heterodimeric complex mafb:cfos bound to2 dna
117 c1ztaA_
not modelled
30.6
16
PDB header: dna-binding motifChain: A: PDB Molecule: leucine zipper monomer;PDBTitle: the solution structure of a leucine-zipper motif peptide
118 c1dh3A_
not modelled
30.2
10
PDB header: transcription/dnaChain: A: PDB Molecule: transcription factor creb;PDBTitle: crystal structure of a creb bzip-cre complex reveals the2 basis for creb faimly selective dimerization and dna3 binding
119 c2gd7B_
not modelled
30.1
21
PDB header: transcriptionChain: B: PDB Molecule: hexim1 protein;PDBTitle: the structure of the cyclin t-binding domain of hexim12 reveals the molecular basis for regulation of3 transcription elongation
120 c4dznC_
not modelled
28.9
19
PDB header: de novo proteinChain: C: PDB Molecule: coiled-coil peptide cc-pil;PDBTitle: a de novo designed coiled coil cc-pil