| 1 |
|
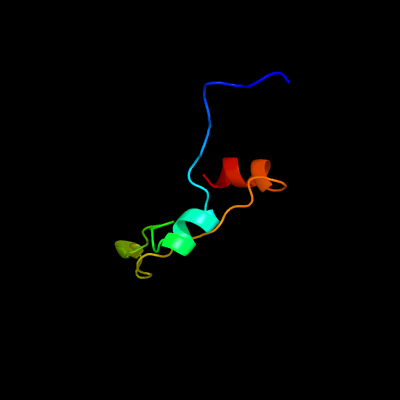
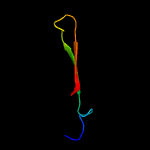
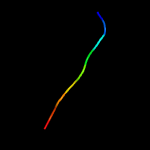
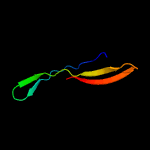
PDB 1xwn chain A domain 1
Region: 21 - 71
Aligned: 40
Modelled: 51
Confidence: 38.6%
Identity: 25%
Fold: Cyclophilin-like
Superfamily: Cyclophilin-like
Family: Cyclophilin (peptidylprolyl isomerase)
Phyre2
| 2 |
|
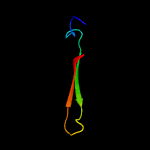
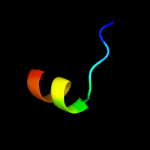
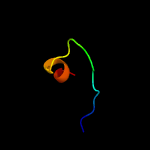
PDB 2y9x chain G
Region: 33 - 60
Aligned: 28
Modelled: 28
Confidence: 23.9%
Identity: 39%
PDB header:oxidoreductase
Chain: G: PDB Molecule:lectin-like fold protein;
PDBTitle: crystal structure of ppo3, a tyrosinase from agaricus bisporus, in2 deoxy-form that contains additional unknown lectin-like subunit,3 with inhibitor tropolone
Phyre2
| 3 |
|
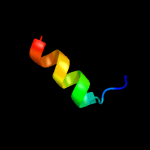
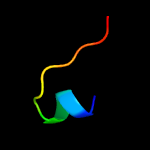
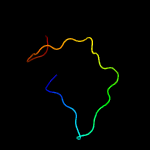
PDB 2w0p chain A domain 1
Region: 3 - 63
Aligned: 61
Modelled: 61
Confidence: 19.3%
Identity: 26%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: Filamin repeat (rod domain)
Phyre2
| 4 |
|
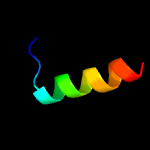
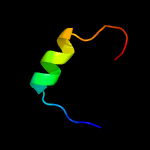
PDB 2c4r chain L
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 14.3%
Identity: 40%
PDB header:hydrolase
Chain: L: PDB Molecule:ribonuclease e;
PDBTitle: catalytic domain of e. coli rnase e
Phyre2
| 5 |
|
PDB 1dwk chain A domain 2
Region: 29 - 59
Aligned: 31
Modelled: 31
Confidence: 11.2%
Identity: 32%
Fold: Cyanase C-terminal domain
Superfamily: Cyanase C-terminal domain
Family: Cyanase C-terminal domain
Phyre2
| 6 |
|
PDB 2iv1 chain J
Region: 29 - 59
Aligned: 31
Modelled: 31
Confidence: 9.8%
Identity: 32%
PDB header:lyase
Chain: J: PDB Molecule:cyanate hydratase;
PDBTitle: site directed mutagenesis of key residues involved in the catalytic2 mechanism of cyanase
Phyre2
| 7 |
|
PDB 5y88 chain S
Region: 25 - 32
Aligned: 8
Modelled: 8
Confidence: 9.3%
Identity: 75%
PDB header:splicing
Chain: S: PDB Molecule:pre-mrna-processing factor 17;
PDBTitle: cryo-em structure of the intron-lariat spliceosome ready for2 disassembly from s.cerevisiae at 3.5 angstrom
Phyre2
| 8 |
|
PDB 3obh chain A
Region: 56 - 71
Aligned: 16
Modelled: 16
Confidence: 8.2%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: x-ray crystal structure of protein sp_0782 (7-79) from streptococcus2 pneumoniae. northeast structural genomics consortium target spr104
Phyre2
| 9 |
|
PDB 2l3a chain A
Region: 56 - 73
Aligned: 18
Modelled: 18
Confidence: 7.8%
Identity: 28%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of homodimer protein sp_0782 (7-79) from2 streptococcus pneumoniae northeast structural genomics consortium3 target spr104 .
Phyre2
| 10 |
|
PDB 2jor chain A
Region: 30 - 38
Aligned: 9
Modelled: 9
Confidence: 6.5%
Identity: 67%
PDB header:blood clotting
Chain: A: PDB Molecule:fibrinogen alpha chain;
PDBTitle: nmr solution structure, stability, and interaction of the2 recombinant bovine fibrinogen alphac-domain fragment
Phyre2
| 11 |
|
PDB 5t5i chain L
Region: 55 - 67
Aligned: 13
Modelled: 13
Confidence: 6.1%
Identity: 23%
PDB header:oxidoreductase
Chain: L: PDB Molecule:tungsten formylmethanofuran dehydrogenase subunit fwdd;
PDBTitle: tungsten-containing formylmethanofuran dehydrogenase from2 methanothermobacter wolfeii, orthorhombic form at 1.9 a
Phyre2
| 12 |
|
PDB 1xdp chain A domain 4
Region: 4 - 16
Aligned: 13
Modelled: 13
Confidence: 6.0%
Identity: 31%
Fold: Phospholipase D/nuclease
Superfamily: Phospholipase D/nuclease
Family: Polyphosphate kinase C-terminal domain
Phyre2
| 13 |
|
PDB 2kke chain A
Region: 55 - 75
Aligned: 21
Modelled: 21
Confidence: 5.7%
Identity: 48%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of a dimeric protein of unknown2 function from methanobacterium thermoautotrophicum,3 northeast structural genomics consortium target tr5
Phyre2
| 14 |
|
PDB 1bvu chain A domain 2
Region: 47 - 67
Aligned: 21
Modelled: 21
Confidence: 5.6%
Identity: 38%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Aminoacid dehydrogenases
Phyre2
| 15 |
|
PDB 4kqz chain A
Region: 47 - 55
Aligned: 9
Modelled: 9
Confidence: 5.6%
Identity: 89%
PDB header:viral protein
Chain: A: PDB Molecule:s protein;
PDBTitle: structure of the receptor binding domain (rbd) of mers-cov spike
Phyre2
| 16 |
|
PDB 2otb chain B
Region: 18 - 59
Aligned: 35
Modelled: 42
Confidence: 5.5%
Identity: 37%
PDB header:fluorescent protein
Chain: B: PDB Molecule:gfp-like fluorescent chromoprotein cfp484;
PDBTitle: crystal structure of a monomeric cyan fluorescent protein in the2 fluorescent state
Phyre2
| 17 |
|
PDB 1gtm chain A domain 2
Region: 47 - 66
Aligned: 20
Modelled: 20
Confidence: 5.5%
Identity: 40%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Aminoacid dehydrogenases
Phyre2
| 18 |
|
PDB 1kqf chain A domain 1
Region: 55 - 67
Aligned: 13
Modelled: 13
Confidence: 5.4%
Identity: 46%
Fold: Double psi beta-barrel
Superfamily: ADC-like
Family: Formate dehydrogenase/DMSO reductase, C-terminal domain
Phyre2
| 19 |
|
PDB 2kke chain B
Region: 55 - 75
Aligned: 21
Modelled: 21
Confidence: 5.2%
Identity: 48%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of a dimeric protein of unknown2 function from methanobacterium thermoautotrophicum,3 northeast structural genomics consortium target tr5
Phyre2
| 20 |
|
PDB 2qdr chain A
Region: 2 - 32
Aligned: 31
Modelled: 31
Confidence: 5.1%
Identity: 29%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative dioxygenase (npun_f5605) from nostoc2 punctiforme pcc 73102 at 2.60 a resolution
Phyre2