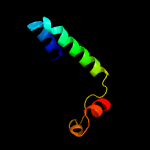
1 c2mi2A_
99.9
23
PDB header: transport proteinChain: A: PDB Molecule: sec-independent protein translocase protein tatb;PDBTitle: solution structure of the e. coli tatb protein in dpc micelles
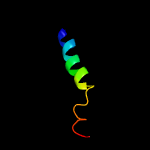
2 c2l16A_
99.8
25
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
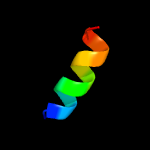
3 c2lzsE_
99.7
13
PDB header: protein transportChain: E: PDB Molecule: sec-independent protein translocase protein tata;PDBTitle: tata oligomer
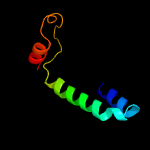
4 c6o7ua_
50.2
18
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
5 c5i1mV_
47.5
17
PDB header: membrane proteinChain: V: PDB Molecule: v-type proton atpase subunit a, vacuolar isoform;PDBTitle: yeast v-atpase average of densities, a subunit segment
6 d1ohua_
39.8
18
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death
7 c6o7xa_
35.0
32
PDB header: membrane proteinChain: A: PDB Molecule: vacuolar atp synthase catalytic subunit a;PDBTitle: saccharomyces cerevisiae v-atpase stv1-v1vo state 3
8 c3a0hJ_
28.9
54
PDB header: electron transportChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: crystal structure of i-substituted photosystem ii complex
9 d2axtj1
24.9
54
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like
10 c3a0hi_
23.2
38
PDB header: electron transportChain: I: PDB Molecule: photosystem ii reaction center protein i;PDBTitle: crystal structure of i-substituted photosystem ii complex
11 d2axti1
23.2
38
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like
12 c5xnmj_
23.1
31
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
13 c3msqC_
22.1
11
PDB header: biosynthetic proteinChain: C: PDB Molecule: putative ubiquinone biosynthesis protein;PDBTitle: crystal structure of a putative ubiquinone biosynthesis protein2 (npun02000094) from nostoc punctiforme pcc 73102 at 2.85 a resolution
14 c3jcuI_
21.4
31
PDB header: membrane proteinChain: I: PDB Molecule: protein photosystem ii reaction center protein i;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
15 c3jcuj_
20.9
38
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
16 c4d18G_
20.1
13
PDB header: signaling proteinChain: G: PDB Molecule: cop9 signalosome complex subunit 7a;PDBTitle: crystal structure of the cop9 signalosome
17 c2voyH_
15.0
28
PDB header: hydrolaseChain: H: PDB Molecule: sarcoplasmic/endoplasmic reticulum calcium atpase 1;PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
18 c4pv1H_
13.6
46
PDB header: electron transport/inhibitorChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: cytochrome b6f structure from m. laminosus with the quinone analog2 inhibitor stigmatellin
19 c4i7zH_
13.6
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of cytochrome b6f in dopg, with disordered rieske2 iron-sulfur protein soluble domain
20 c3kb4D_
13.4
7
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr8543 protein;PDBTitle: crystal structure of the alr8543 protein in complex with2 geranylgeranyl monophosphate and magnesium ion from nostoc sp. pcc3 7120, northeast structural genomics consortium target nsr141
21 c2e76H_
not modelled
13.3
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
22 c2e75H_
not modelled
13.3
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
23 c2e74H_
not modelled
13.3
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
24 c4h13H_
not modelled
13.3
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex from mastigocladus2 laminosus with tds
25 c4h0lH_
not modelled
13.3
46
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: cytochrome b6f complex crystal structure from mastigocladus laminosus2 with n-side inhibitor nqno
26 c5uulB_
not modelled
12.5
19
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-binding component 3;PDBTitle: human bfl-1 in complex with puma bh3
27 c4pj0L_
not modelled
11.1
14
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
28 c4pj0l_
not modelled
11.1
14
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
29 c4rvyL_
not modelled
10.7
14
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
30 c4tniL_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
31 c4rvyl_
not modelled
10.7
14
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
32 c3a0bl_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
33 c3a0hl_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
34 c4tnkl_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
35 c5e7cL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
36 c3prrL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
37 c4tnhL_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
38 c4fbyL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: fs x-ray diffraction of photosystem ii
39 c2axtl_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
40 c5e7cl_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
41 c3a0bL_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
42 c4tnjL_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
43 c4tnhl_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
44 c4fbyd_
not modelled
10.7
14
PDB header: photosynthesisChain: D: PDB Molecule: photosystem ii d2 protein;PDBTitle: fs x-ray diffraction of photosystem ii
45 c3arcL_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
46 c2axtL_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
47 c4tnil_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
48 d2axtl1
not modelled
10.7
14
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like
49 c3prqL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
50 c4tnkL_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
51 c3a0hL_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
52 c4tnjl_
not modelled
10.7
14
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
53 c4ixrL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
54 c3wu2L_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
55 c4ixqL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
56 c4il6l_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
57 c4il6L_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
58 c3wu2l_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
59 c3bz2L_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 2 of 2). this2 file contains second monomer of psii dimer
60 c4ixrl_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
61 c4ixql_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
62 c3kziL_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
63 c3bz1L_
not modelled
10.7
14
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 1 of 2). this2 file contains first monomer of psii dimer
64 c1s5ll_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
65 c4ub6l_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
66 c4ub8L_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
67 c1s5lL_
not modelled
10.7
14
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
68 c4ub8l_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
69 c4ub6L_
not modelled
10.7
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
70 c2bzwB_
not modelled
9.5
16
PDB header: transcriptionChain: B: PDB Molecule: bcl2-antagonist of cell death;PDBTitle: the crystal structure of bcl-xl in complex with full-length bad
71 c3arcl_
not modelled
9.2
14
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
72 c1kv4A_
not modelled
8.6
17
PDB header: antibioticChain: A: PDB Molecule: moricin;PDBTitle: solution structure of antibacterial peptide (moricin)
73 c2o60B_
not modelled
8.4
26
PDB header: metal binding proteinChain: B: PDB Molecule: peptide corresponding to calmodulin binding domain ofPDBTitle: calmodulin bound to peptide from neuronal nitric oxide synthase
74 c4nf9A_
not modelled
7.8
11
PDB header: cell cycleChain: A: PDB Molecule: protein casc5;PDBTitle: structure of the knl1/nsl1 complex
75 c2jpxA_
not modelled
7.7
50
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: a18h vpu tm structure in lipid bilayers
76 c6dkmB_
not modelled
7.5
12
PDB header: de novo proteinChain: B: PDB Molecule: dhd131_b;PDBTitle: dhd131
77 c2jr8A_
not modelled
7.5
17
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide moricin;PDBTitle: solution structure of manduca sexta moricin
78 c2zjsE_
not modelled
7.5
13
PDB header: protein transport/immune systemChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: crystal structure of secye translocon from thermus thermophilus with a2 fab fragment
79 c5ireD_
not modelled
7.4
25
PDB header: virusChain: D: PDB Molecule: m protein;PDBTitle: the cryo-em structure of zika virus
80 c2rocB_
not modelled
7.3
17
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-binding component 3;PDBTitle: solution structure of mcl-1 complexed with puma
81 c3ci9B_
not modelled
6.7
12
PDB header: transcriptionChain: B: PDB Molecule: heat shock factor-binding protein 1;PDBTitle: crystal structure of the human hsbp1
82 c5t42A_
not modelled
6.7
18
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: structure of the ebola virus envelope protein mper/tm domain and its2 interaction with the fusion loop explains their fusion activity
83 c6btmA_
not modelled
6.7
31
PDB header: membrane proteinChain: A: PDB Molecule: alternative complex iii subunit a;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
84 c5xauC_
not modelled
6.6
9
PDB header: cell adhesionChain: C: PDB Molecule: laminin subunit gamma-1;PDBTitle: crystal structure of integrin binding fragment of laminin-511
85 c5wdaL_
not modelled
6.6
36
PDB header: protein transportChain: L: PDB Molecule: general secretion pathway protein g;PDBTitle: structure of the pulg pseudopilus
86 d2cqqa1
not modelled
6.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain
87 c5j10A_
not modelled
6.4
11
PDB header: de novo proteinChain: A: PDB Molecule: peptide design 2l4hc2_24;PDBTitle: de novo design of protein homo-oligomers with modular hydrogen bond2 network-mediated specificity
88 c5wsnD_
not modelled
6.3
25
PDB header: virusChain: D: PDB Molecule: m protein;PDBTitle: structure of japanese encephalitis virus
89 c1p58E_
not modelled
5.9
33
PDB header: virusChain: E: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
90 c1p58F_
not modelled
5.9
33
PDB header: virusChain: F: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
91 c6cfzC_
not modelled
5.8
26
PDB header: nuclear proteinChain: C: PDB Molecule: dad2;PDBTitle: structure of the dash/dam1 complex shows its role at the yeast2 kinetochore-microtubule interface
92 c3mwgA_
not modelled
5.8
9
PDB header: transport proteinChain: A: PDB Molecule: iron-regulated abc transporter siderophore-binding proteinPDBTitle: crystal structure of staphylococcus aureus sira
93 c4hnjC_
not modelled
5.7
22
PDB header: apoptosis/protein bindingChain: C: PDB Molecule: bcl-2-binding component 3;PDBTitle: crystallographic structure of bcl-xl domain-swapped dimer in complex2 with puma bh3 peptide at 2.9a resolution
94 c3t0yA_
not modelled
5.7
8
PDB header: transcription regulator/protein bindingChain: A: PDB Molecule: response regulator;PDBTitle: structure of the phyr anti-anti-sigma domain bound to the anti-sigma2 factor, nepr
95 c6fkip_
not modelled
5.7
4
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
96 c6fo1G_
not modelled
5.4
21
PDB header: chaperoneChain: G: PDB Molecule: rna polymerase ii-associated protein 3;PDBTitle: human r2tp subcomplex containing 1 ruvbl1-ruvbl2 hexamer bound to 12 rbd domain from rpap3.
97 c3jcul_
not modelled
5.4
14
PDB header: membrane proteinChain: L: PDB Molecule: protein photosystem ii reaction center protein l;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
98 c3nzlA_
not modelled
5.2
0
PDB header: transcriptionChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: crystal structure of the n-terminal domain of dna-binding protein2 satb1 from homo sapiens, northeast structural genomics consortium3 target hr4435b
99 c3zbhC_
not modelled
5.2
16
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i