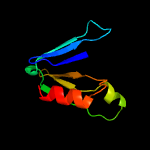
1 d2gcla1
74.2
19
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: SSRP1-like
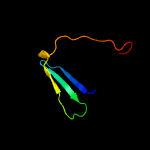
2 c3fssA_
51.4
8
PDB header: chaperoneChain: A: PDB Molecule: histone chaperone rtt106;PDBTitle: structure of the tandem ph domains of rtt106
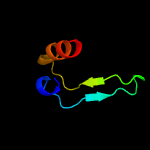
3 c6c4vA_
49.9
18
PDB header: transport proteinChain: A: PDB Molecule: polyketide synthase pks13;PDBTitle: 1.9 angstrom resolution crystal structure of acyl carrier protein2 domain (residues 1350-1461) of polyketide synthase pks13 from3 mycobacterium tuberculosis
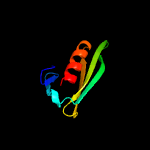
4 c4tyzB_
49.4
11
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the c-terminal domain of an unknown protein from2 leishmania infantum
5 d2hthb1
48.8
7
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: VPS36 N-terminal domain-like
6 d1zj8a1
39.9
16
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
7 c3gypA_
29.5
8
PDB header: chaperoneChain: A: PDB Molecule: histone chaperone rtt106;PDBTitle: rtt106p
8 d1j8ba_
29.4
18
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like
9 d3c7bb2
28.2
11
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like
10 d2v4jb2
22.8
14
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like
11 d2ebfx3
21.9
14
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: PMT C-terminal domain like
12 d2caya1
20.3
11
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: VPS36 N-terminal domain-like
13 c1ybxA_
19.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
14 c5nocA_
17.3
23
PDB header: dna binding proteinChain: A: PDB Molecule: stage 0 sporulation protein j;PDBTitle: solution nmr structure of the c-terminal domain of parb (spo0j)
15 d3dcxa1
16.1
5
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: BPHL domain
16 d1aopa2
15.0
6
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
17 c4n7rD_
12.0
13
PDB header: oxidoreductase/protein bindingChain: D: PDB Molecule: genomic dna, chromosome 3, p1 clone: mxl8;PDBTitle: crystal structure of arabidopsis glutamyl-trna reductase in complex2 with its binding protein
18 c2fwvA_
12.0
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein mtubf_01000852;PDBTitle: crystal structure of rv0813
19 d1zj8a2
11.1
11
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
20 c5x3xq_
10.6
13
PDB header: transport proteinChain: Q: PDB Molecule: uncharacterized protein cbiq;PDBTitle: 2.8a resolution structure of a cobalt energy-coupling factor2 transporter-cbimqo
21 c5bncB_
not modelled
10.2
15
PDB header: heme binding proteinChain: B: PDB Molecule: heme binding protein msmeg_6519;PDBTitle: structure of heme binding protein msmeg_6519 from mycobacterium2 smegmatis
22 d1puga_
not modelled
10.1
20
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like
23 c2d7dB_
not modelled
10.1
32
PDB header: hydrolase/dnaChain: B: PDB Molecule: 40-mer from uvrabc system protein b;PDBTitle: structural insights into the cryptic dna dependent atp-ase2 activity of uvrb
24 c2v4jE_
not modelled
10.0
14
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
25 c3c7bE_
not modelled
9.3
11
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
26 d2akja1
not modelled
8.6
11
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
27 c2v4jA_
not modelled
8.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
28 c3c7bA_
not modelled
7.8
11
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunit alpha;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
29 d1iioa_
not modelled
7.3
22
Fold: EF Hand-likeSuperfamily: Hypothetical protein MTH865Family: Hypothetical protein MTH865
30 c2kw0A_
not modelled
6.6
7
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
31 c6eznH_
not modelled
6.6
17
PDB header: membrane proteinChain: H: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
32 c2kl8A_
not modelled
6.3
16
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like fold2 protein, northeast structural genomics consortium target or15
33 d2dx5a1
not modelled
6.2
13
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: VPS36 N-terminal domain-like
34 c4e1pA_
not modelled
6.1
20
PDB header: dna binding proteinChain: A: PDB Molecule: protein lsr2;PDBTitle: crystal structure of the dimerization domain of lsr2 from2 mycobacterium tuberculosis in the p 1 21 1 space group
35 c4e1rA_
not modelled
6.1
20
PDB header: dna binding proteinChain: A: PDB Molecule: protein lsr2;PDBTitle: crystal structure of the dimerization domain of lsr2 from2 mycobacterium tuberculosis in the p 31 2 1 space group
36 c2n2aA_
not modelled
6.0
13
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
37 c4ev6E_
not modelled
5.7
13
PDB header: metal transportChain: E: PDB Molecule: magnesium transport protein cora;PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
38 c2hl7A_
not modelled
5.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
39 c5h92A_
not modelled
5.5
14
PDB header: oxidoreductase/electron transportChain: A: PDB Molecule: sulfite reductase [ferredoxin], chloroplastic;PDBTitle: crystal structure of the complex between maize sulfite reductase and2 ferredoxin in the form-3 crystal