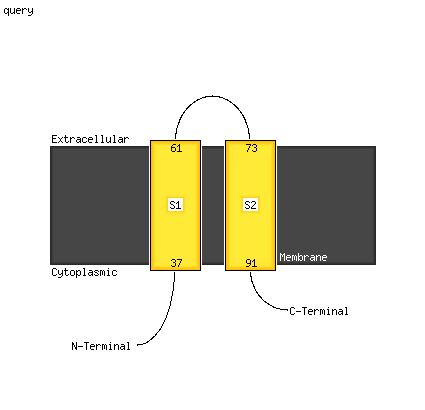
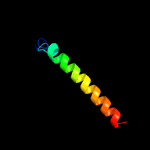
1 c6cfwA_
51.5
18
PDB header: membrane proteinChain: A: PDB Molecule: monovalent cation/h+ antiporter subunit e;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
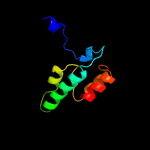
2 c4cbfE_
44.5
23
PDB header: virusChain: E: PDB Molecule: envelope protein e;PDBTitle: near-atomic resolution cryo-em structure of dengue serotype 4 virus
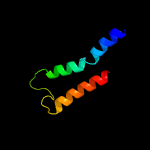
3 c5ireA_
34.6
27
PDB header: virusChain: A: PDB Molecule: e protein;PDBTitle: the cryo-em structure of zika virus
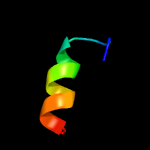
4 c5o6vC_
32.3
32
PDB header: virusChain: C: PDB Molecule: envelope protein;PDBTitle: the cryo-em structure of tick-borne encephalitis virus complexed with2 fab fragment of neutralizing antibody 19/1786
5 c6bwiA_
28.0
8
PDB header: membrane proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: 3.7 angstrom cryoem structure of full length human trpm4
6 c5wsnC_
27.8
23
PDB header: virusChain: C: PDB Molecule: e protein;PDBTitle: structure of japanese encephalitis virus
7 d1k4ta1
26.0
18
Fold: Long alpha-hairpinSuperfamily: Eukaryotic DNA topoisomerase I, dispensable insert domainFamily: Eukaryotic DNA topoisomerase I, dispensable insert domain
8 c4b03A_
20.4
32
PDB header: virusChain: A: PDB Molecule: dengue virus 1 e protein;PDBTitle: 6a electron cryomicroscopy structure of immature dengue virus serotype2 1
9 c6co7C_
18.2
9
PDB header: membrane proteinChain: C: PDB Molecule: predicted protein;PDBTitle: structure of the nvtrpm2 channel in complex with ca2+
10 c4ltpC_
17.9
16
PDB header: transport proteinChain: C: PDB Molecule: ion transport protein;PDBTitle: bacterial sodium channel in high calcium, i222 space group, crystal 2
11 c1p58C_
17.1
24
PDB header: virusChain: C: PDB Molecule: major envelope protein e;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
12 c6rdu9_
17.1
17
PDB header: proton transportChain: 9: PDB Molecule: asa-9: polytomella f-atp synthase associated subunit 9;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 monomer-masked refinement
13 c2mxbA_
16.9
22
PDB header: membrane proteinChain: A: PDB Molecule: erythropoietin receptor;PDBTitle: structure of the transmembrane domain of the mouse erythropoietin2 receptor
14 c2w2eA_
15.7
17
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin pip2-7 7;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin, aqy1, in a2 closed conformation at ph 3.5
15 c4ev6E_
15.6
11
PDB header: metal transportChain: E: PDB Molecule: magnesium transport protein cora;PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
16 c3m9bK_
15.4
19
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
17 d1dfma_
15.2
40
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BglII
18 c2m6xC_
14.0
20
PDB header: membrane proteinChain: C: PDB Molecule: p7;PDBTitle: structure of the p7 channel of hepatitis c virus, genotype 5a
19 c2kn8A_
11.4
20
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: dna cleavage and packaging protein large subunit, ul89;PDBTitle: nmr structure of the c-terminal domain of pul89
20 c6bwiB_
10.5
16
PDB header: membrane proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: 3.7 angstrom cryoem structure of full length human trpm4
21 c6bwiD_
not modelled
10.5
16
PDB header: membrane proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: 3.7 angstrom cryoem structure of full length human trpm4
22 c6bwiC_
not modelled
10.5
16
PDB header: membrane proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: 3.7 angstrom cryoem structure of full length human trpm4
23 c6bcqB_
not modelled
10.3
14
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of trpm4 in atp bound state with long coiled coil at2 3.3 angstrom resolution
24 c1ci6A_
not modelled
10.0
20
PDB header: transcriptionChain: A: PDB Molecule: transcription factor atf-4;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
25 c5zx5D_
not modelled
9.6
13
PDB header: transferaseChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: 3.3 angstrom structure of mouse trpm7 with edta
26 d2i7pa1
not modelled
9.6
56
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like
27 c2mv6A_
not modelled
9.2
29
PDB header: membrane proteinChain: A: PDB Molecule: erythropoietin receptor;PDBTitle: solution structure of the transmembrane domain and the juxta-membrane2 domain of the erythropoietin receptor in micelles
28 c4uicA_
not modelled
8.4
23
PDB header: sugar binding proteinChain: A: PDB Molecule: surface layer protein;PDBTitle: crystal structure of the s-layer protein rsbsc(31-844)
29 c5fv8A_
not modelled
8.4
33
PDB header: structural proteinChain: A: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
30 c1d7mA_
not modelled
8.1
30
PDB header: contractile proteinChain: A: PDB Molecule: cortexillin i;PDBTitle: coiled-coil dimerization domain from cortexillin i
31 d2i7na2
not modelled
8.0
56
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like
32 c3hd6A_
not modelled
8.0
17
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
33 c2bbjB_
not modelled
7.9
10
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
34 c2lifA_
not modelled
7.7
38
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: core protein p21;PDBTitle: solution structure of kkgf
35 c2rsmA_
not modelled
7.6
26
PDB header: translationChain: A: PDB Molecule: probable peptide chain release factor c12orf65 homolog,PDBTitle: solution structure and sirna-mediated knockdown analysis of the2 mitochondrial disease-related protein c12orf65 (ict2)
36 c5fv8B_
not modelled
7.2
35
PDB header: structural proteinChain: B: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
37 c3a5tB_
not modelled
7.1
22
PDB header: transcription regulator/dnaChain: B: PDB Molecule: transcription factor mafg;PDBTitle: crystal structure of mafg-dna complex
38 c3vjjA_
not modelled
6.9
50
PDB header: viral proteinChain: A: PDB Molecule: p9-1;PDBTitle: crystal structure analysis of the p9-1
39 d2f9wa2
not modelled
6.6
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: CoaX-like
40 c2js5B_
not modelled
6.5
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of protein q60c73_metca. northeast structural2 genomics consortium target mcr1
41 c3vviB_
not modelled
6.3
40
PDB header: transport proteinChain: B: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
42 c3vviF_
not modelled
6.3
40
PDB header: transport proteinChain: F: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
43 c3vviC_
not modelled
6.3
40
PDB header: transport proteinChain: C: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
44 c3vviG_
not modelled
6.3
40
PDB header: transport proteinChain: G: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
45 c3vviA_
not modelled
6.3
40
PDB header: transport proteinChain: A: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
46 c3vviE_
not modelled
6.3
40
PDB header: transport proteinChain: E: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
47 c3vviH_
not modelled
6.3
40
PDB header: transport proteinChain: H: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
48 c3vviD_
not modelled
6.3
40
PDB header: transport proteinChain: D: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
49 d2a65a1
not modelled
6.2
12
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like
50 c2kl5A_
not modelled
6.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yutd;PDBTitle: solution nmr structure of protein yutd from b.subtilis, northeast2 structural genomics consortium target sr232
51 c6b2zA_
not modelled
6.1
29
PDB header: membrane proteinChain: A: PDB Molecule: atp synthase protein 8;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
52 c6b2zL_
not modelled
6.1
29
PDB header: membrane proteinChain: L: PDB Molecule: atp synthase protein 8;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
53 c5gpdB_
not modelled
6.0
33
PDB header: dna binding proteinChain: B: PDB Molecule: sterol regulatory element-binding protein 1;PDBTitle: crystal structure of the binding domain of srebp from fission yeast
54 c5khtD_
not modelled
5.9
32
PDB header: actin-binding proteinChain: D: PDB Molecule: tropomyosin alpha-1 chain,general control protein gcn4;PDBTitle: crystal structure of the n-terminal fragment of tropomyosin isoform2 tpm1.1 at 1.5 a resolution
55 c3iynQ_
not modelled
5.7
17
PDB header: virusChain: Q: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
56 c5nikK_
not modelled
5.6
24
PDB header: transport proteinChain: K: PDB Molecule: macrolide export atp-binding/permease protein macb;PDBTitle: structure of the macab-tolc abc-type tripartite multidrug efflux pump
57 c5b7iA_
not modelled
5.6
10
PDB header: hydrolase/unknown functionChain: A: PDB Molecule: crispr-associated nuclease/helicase cas3 subtype i-f/ypest;PDBTitle: cas3-acrf3 complex
58 c1wq6A_
not modelled
5.5
19
PDB header: oncoproteinChain: A: PDB Molecule: aml1-eto;PDBTitle: the tetramer structure of the nervy homolgy two (nhr2) domain of aml1-2 eto is critical for aml1-eto's activity
59 c5ey2A_
not modelled
5.5
22
PDB header: transcriptionChain: A: PDB Molecule: gtp-sensing transcriptional pleiotropic repressor cody;PDBTitle: crystal structure of cody from bacillus cereus
60 c5ed9A_
not modelled
5.4
28
PDB header: transport proteinChain: A: PDB Molecule: sun domain-containing protein 2;PDBTitle: crystal structure of cc1 of mouse sun2
61 c5l75F_
not modelled
5.2
16
PDB header: transport proteinChain: F: PDB Molecule: fig000988: predicted permease;PDBTitle: a protein structure
62 c6c9aB_
not modelled
5.2
14
PDB header: membrane proteinChain: B: PDB Molecule: two pore calcium channel protein 1;PDBTitle: cryo-em structure of mouse tpc1 channel in the ptdins(3,5)p2-bound2 state
63 c6e3yE_
not modelled
5.1
10
PDB header: signaling proteinChain: E: PDB Molecule: receptor activity-modifying protein 1;PDBTitle: cryo-em structure of the active, gs-protein complexed, human cgrp2 receptor
64 d2vbua1
not modelled
5.1
40
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like
65 c5aoqA_
not modelled
5.0
63
PDB header: transferaseChain: A: PDB Molecule: torso;PDBTitle: structural basis of neurohormone perception by the receptor tyrosine2 kinase torso
66 d1u7ga_
not modelled
5.0
15
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter