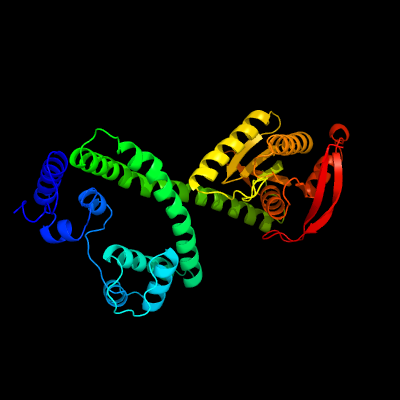
| 1 | c1y10C_
|
|
 |
100.0 |
100 |
PDB header:lyase
Chain: C: PDB Molecule:hypothetical protein rv1264/mt1302;
PDBTitle: mycobacterial adenylyl cyclase rv1264, holoenzyme, inhibited state
|
|
|
|
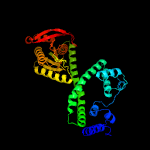
| 2 | c6fhtB_
|
|
 |
100.0 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:bacteriophytochrome,adenylate cyclase;
PDBTitle: crystal structure of an artificial phytochrome regulated2 adenylate/guanylate cyclase in its dark adapted pr form
|
|
|
|
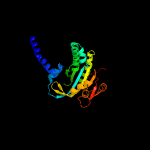
| 3 | c2ev2B_
|
|
 |
100.0 |
100 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein rv1264/mt1302;
PDBTitle: structure of rv1264n, the regulatory domain of the mycobacterial2 adenylyl cylcase rv1264, at ph 8.5
|
|
|
|
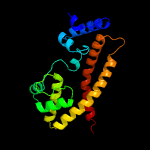
| 4 | c4yusA_
|
|
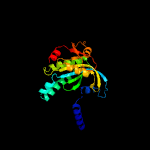 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:family 3 adenylate cyclase;
PDBTitle: crystal structure of photoactivated adenylyl cyclase of a2 cyanobacteriaoscillatoria acuminata in hexagonal form
|
|
|
|
| 5 | d1fx2a_
|
|
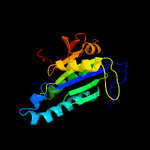 |
100.0 |
22 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
|
|
|
| 6 | d1fx4a_
|
|
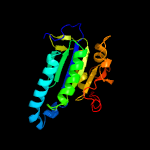 |
100.0 |
23 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
|
|
|
| 7 | c5nbyA_
|
|
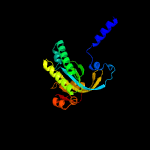 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:beta subunit of photoactivated adenylyl cyclase;
PDBTitle: structure of a bacterial light-regulated adenylyl cylcase
|
|
|
|
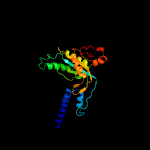
| 8 | c6r4oA_
|
|
 |
100.0 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:adenylate cyclase 9;
PDBTitle: structure of a truncated adenylyl cyclase bound to mant-gtp, forskolin2 and an activated stimulatory galphas protein
|
|
|
|
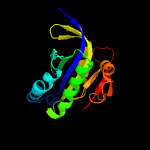
| 9 | c3mr7B_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenylate/guanylate cyclase/hydrolase, alpha/beta fold
PDBTitle: crystal structure of adenylate/guanylate cyclase/hydrolase from2 silicibacter pomeroyi
|
|
|
|
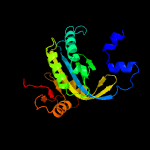
| 10 | c4cllA_
|
|
 |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:adenylate cyclase type 10;
PDBTitle: crystal structure of human soluble adenylyl cyclase in complex with2 bicarbonate
|
|
|
|
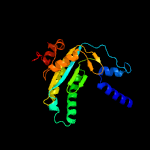
| 11 | c6r4pA_
|
|
 |
100.0 |
21 |
PDB header:membrane protein
Chain: A: PDB Molecule:adenylate cyclase 9;
PDBTitle: structure of a soluble domain of adenylyl cyclase bound to an2 activated stimulatory g protein
|
|
|
|
| 12 | c4wp3E_
|
|
 |
100.0 |
31 |
PDB header:lyase
Chain: E: PDB Molecule:ma1120;
PDBTitle: crystal structure of adenylyl cyclase from mycobacterium avium ma11202 wild type
|
|
|
|
| 13 | c5o5kC_
|
|
 |
100.0 |
28 |
PDB header:membrane protein
Chain: C: PDB Molecule:adenylate cyclase;
PDBTitle: x-ray structure of a bacterial adenylyl cyclase soluble domain
|
|
|
|
| 14 | c1wc6B_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:adenylate cyclase;
PDBTitle: soluble adenylyl cyclase cyac from s. platensis in complex2 with rp-atpalphas in presence of bicarbonate
|
|
|
|
| 15 | c1ybuA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipj;
PDBTitle: mycobacterium tuberculosis adenylyl cyclase rv1900c chd, in complex2 with a substrate analog.
|
|
|
|
| 16 | d1wc1a_
|
|
 |
100.0 |
26 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
|
|
|
| 17 | c3uvjC_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: C: PDB Molecule:guanylate cyclase soluble subunit alpha-3;
PDBTitle: crystal structure of the catalytic domain of the heterodimeric human2 soluble guanylate cyclase 1.
|
|
|
|
| 18 | c1cjkA_
|
|
 |
100.0 |
23 |
PDB header:lyase/lyase/signaling protein
Chain: A: PDB Molecule:adenylate cyclase, type v;
PDBTitle: complex of gs-alpha with the catalytic domains of mammalian adenylyl2 cyclase: complex with adenosine 5'-(alpha thio)-triphosphate (rp),3 mg, and mn
|
|
|
|
| 19 | c3r5gB_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:cyab;
PDBTitle: crystal structure of the adenylyl cyclase cyab from p. aeruginosa
|
|
|
|
| 20 | c2w01C_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:adenylate cyclase;
PDBTitle: crystal structure of the guanylyl cyclase cya2
|
|
|
|
| 21 | d1azsa_ |
|
not modelled |
100.0 |
21 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
|
|
| 22 | c3et6A_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:soluble guanylyl cyclase beta;
PDBTitle: the crystal structure of the catalytic domain of a eukaryotic2 guanylate cyclase
|
|
|
| 23 | c2wz1B_ |
|
not modelled |
100.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:guanylate cyclase soluble subunit beta-1;
PDBTitle: structure of the catalytic domain of human soluble guanylate cyclase 12 beta 3.
|
|
|
| 24 | d1azsb_ |
|
not modelled |
100.0 |
27 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:Adenylyl and guanylyl cyclase catalytic domain |
|
|
| 25 | c1yk9A_ |
|
not modelled |
100.0 |
31 |
PDB header:lyase
Chain: A: PDB Molecule:adenylate cyclase;
PDBTitle: crystal structure of a mutant form of the mycobacterial2 adenylyl cyclase rv1625c
|
|
|
| 26 | c6aoaA_ |
|
not modelled |
99.9 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:bacterio-rhodopsin/guanylyl cyclase 1 fusion protein;
PDBTitle: monomeric crystal structure of the e497/c566d double mutant of the2 guanylyl cyclase domain of the rhogc fusion protein from the aquatic3 fungus blastocladiella emersonii
|
|
|
| 27 | c4zmuD_ |
|
not modelled |
96.2 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:diguanylate cyclase;
PDBTitle: dcsbis, a diguanylate cyclase from pseudomonas aeruginosa
|
|
|
| 28 | c5llxB_ |
|
not modelled |
95.7 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:diguanylate cyclase (ggdef) domain-containing protein;
PDBTitle: bacteriophytochrome activated diguanylyl cyclase from idiomarina2 species a28l with gtp bound
|
|
|
| 29 | c5c8eC_ |
|
not modelled |
95.5 |
14 |
PDB header:transcription regulator/dna
Chain: C: PDB Molecule:light-dependent transcriptional regulator carh;
PDBTitle: crystal structure of thermus thermophilus carh bound to2 adenosylcobalamin and a 26-bp dna segment
|
|
|
| 30 | c2qv6D_ |
|
not modelled |
95.2 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase iii;
PDBTitle: gtp cyclohydrolase iii from m. jannaschii (mj0145)2 complexed with gtp and metal ions
|
|
|
| 31 | c1w25B_ |
|
not modelled |
94.4 |
12 |
PDB header:signaling protein
Chain: B: PDB Molecule:stalked-cell differentiation controlling protein;
PDBTitle: response regulator pled in complex with c-digmp
|
|
|
| 32 | c4r4eA_ |
|
not modelled |
94.1 |
10 |
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:hth-type transcriptional regulator glnr;
PDBTitle: structure of glnr-dna complex
|
|
|
| 33 | c3ezuA_ |
|
not modelled |
93.8 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:ggdef domain protein;
PDBTitle: crystal structure of multidomain protein of unknown function with2 ggdef-domain (np_951600.1) from geobacter sulfurreducens at 1.95 a3 resolution
|
|
|
| 34 | c4r24B_ |
|
not modelled |
93.4 |
15 |
PDB header:transcrption/dna
Chain: B: PDB Molecule:hth-type transcriptional regulator tnra;
PDBTitle: complete dissection of b. subtilis nitrogen homeostatic circuitry
|
|
|
| 35 | c5af3A_ |
|
not modelled |
93.2 |
11 |
PDB header:dna binding
Chain: A: PDB Molecule:vapbc49;
PDBTitle: x-ray crystal structure of rv2018 from mycobacterium tuberculosis
|
|
|
| 36 | c3hh0C_ |
|
not modelled |
91.6 |
15 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulator, merr family;
PDBTitle: crystal structure of a transcriptional regulator, merr family from2 bacillus cereus
|
|
|
| 37 | c5ydcA_ |
|
not modelled |
91.2 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:uncharacterized hth-type transcriptional regulator rv1828;
PDBTitle: crystal structure of mercury soaked c-terminal domain of rv1828 from2 mycobacterium tuberculosis
|
|
|
| 38 | c6d9mA_ |
|
not modelled |
91.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:fusion protein of endolysin,response receiver sensor
PDBTitle: t4-lysozyme fusion to geobacter ggdef
|
|
|
| 39 | c3ignA_ |
|
not modelled |
89.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:diguanylate cyclase;
PDBTitle: crystal structure of the ggdef domain from marinobacter aquaeolei2 diguanylate cyclase complexed with c-di-gmp - northeast structural3 genomics consortium target mqr89a
|
|
|
| 40 | c2zhhA_ |
|
not modelled |
89.4 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:redox-sensitive transcriptional activator soxr;
PDBTitle: crystal structure of soxr
|
|
|
| 41 | c4h54B_ |
|
not modelled |
89.4 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:diguanylate cyclase ydeh;
PDBTitle: crystal structure of the diguanylate cyclase dgcz
|
|
|
| 42 | c5i44E_ |
|
not modelled |
88.5 |
13 |
PDB header:dna binding protein/dna
Chain: E: PDB Molecule:chromosome-anchoring protein raca;
PDBTitle: structure of raca-dna complex; p21 form
|
|
|
| 43 | c3mtkA_ |
|
not modelled |
88.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:diguanylate cyclase/phosphodiesterase;
PDBTitle: x-ray structure of diguanylate cyclase/phosphodiesterase from2 caldicellulosiruptor saccharolyticus, northeast structural genomics3 consortium target clr27c
|
|
|
| 44 | c5crlA_ |
|
not modelled |
88.0 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:mercuric resistance operon regulatory protein;
PDBTitle: crystal structure of the transcription activator tn501 merr in complex2 with mercury (ii)
|
|
|
| 45 | d1r8da_ |
|
not modelled |
87.4 |
18 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:DNA-binding N-terminal domain of transcription activators |
|
|
| 46 | c5gpeB_ |
|
not modelled |
85.7 |
7 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, merr-family;
PDBTitle: crystal structure of the transcription regulator pbrr691 from2 ralstonia metallidurans ch34 in complex with lead(ii)
|
|
|
| 47 | d1r8ea1 |
|
not modelled |
85.5 |
14 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:DNA-binding N-terminal domain of transcription activators |
|
|
| 48 | c2vz4A_ |
|
not modelled |
84.0 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional activator tipa;
PDBTitle: the n-terminal domain of merr-like protein tipal bound to promoter dna
|
|
|
| 49 | c4ymeA_ |
|
not modelled |
83.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:sensory box/ggdef family protein;
PDBTitle: crystal structure of a sensory box/ggdef family protein (cc_0091) from2 caulobacter crescentus cb15 at 1.40 a resolution (psi community3 target, shapiro)
|
|
|
| 50 | c6eibC_ |
|
not modelled |
82.8 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:sensory box/ggdef family protein;
PDBTitle: structure of the active ggeef domain of a diguanylate cyclase from2 vibrio cholerae.
|
|
|
| 51 | c4zvhB_ |
|
not modelled |
82.3 |
15 |
PDB header:signaling protein
Chain: B: PDB Molecule:diguanylate cyclase dosc;
PDBTitle: crystal structure of ggdef domain of the e. coli dosc - form iv
|
|
|
| 52 | c3hvwA_ |
|
not modelled |
81.4 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:diguanylate-cyclase (dgc);
PDBTitle: crystal structure of the ggdef domain of the pa2567 protein from2 pseudomonas aeruginosa, northeast structural genomics consortium3 target par365c
|
|
|
| 53 | c2jmlA_ |
|
not modelled |
80.3 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:dna binding domain/transcriptional regulator;
PDBTitle: solution structure of the n-terminal domain of cara repressor
|
|
|
| 54 | c3hvaA_ |
|
not modelled |
80.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:protein fimx;
PDBTitle: crystal structure of fimx ggdef domain from pseudomonas aeruginosa
|
|
|
| 55 | c4iobA_ |
|
not modelled |
79.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:diguanylate cyclase tpbb;
PDBTitle: crystal structure of the ggdef domain of pa1120 (yfin or tpbb) from2 pseudomonas aeruginosa at 2.7 ang.
|
|
|
| 56 | c4wxoA_ |
|
not modelled |
79.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: sadc (300-487) from pseudomonas aeruginosa pao1
|
|
|
| 57 | c3ucsB_ |
|
not modelled |
77.4 |
19 |
PDB header:chaperone
Chain: B: PDB Molecule:chaperone-modulator protein cbpm;
PDBTitle: crystal structure of the complex between cbpa j-domain and cbpm
|
|
|
| 58 | c3gp4B_ |
|
not modelled |
77.4 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, merr family;
PDBTitle: crystal structure of putative merr family transcriptional regulator2 from listeria monocytogenes
|
|
|
| 59 | c3i5aA_ |
|
not modelled |
76.1 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator/ggdef domain protein;
PDBTitle: crystal structure of full-length wpsr from pseudomonas syringae
|
|
|
| 60 | c4zmmB_ |
|
not modelled |
75.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:diguanylate cyclase;
PDBTitle: ggdef domain of dcsbis complexed with c-di-gmp
|
|
|
| 61 | c3breA_ |
|
not modelled |
74.8 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:probable two-component response regulator;
PDBTitle: crystal structure of p.aeruginosa pa3702
|
|
|
| 62 | c4euvA_ |
|
not modelled |
72.8 |
8 |
PDB header:signaling protein
Chain: A: PDB Molecule:peld;
PDBTitle: crystal structure of peld 158-ct from pseudomonas aeruginosa pao1, in2 complex with c-di-gmp, form 1
|
|
|
| 63 | d1q06a_ |
|
not modelled |
70.7 |
10 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:DNA-binding N-terminal domain of transcription activators |
|
|
| 64 | c3gpvA_ |
|
not modelled |
70.5 |
6 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, merr family;
PDBTitle: crystal structure of a transcriptional regulator, merr2 family from bacillus thuringiensis
|
|
|
| 65 | c3iclA_ |
|
not modelled |
67.4 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:eal/ggdef domain protein;
PDBTitle: x-ray structure of protein (eal/ggdef domain protein) from2 m.capsulatus, northeast structural genomics consortium target mcr174c
|
|
|
| 66 | c6amaO_ |
|
not modelled |
62.7 |
17 |
PDB header:dna binding protein/dna
Chain: O: PDB Molecule:putative dna-binding protein;
PDBTitle: structure of s. coelicolor/s. venezuelae bldc-smea-ssfa complex to2 3.09 angstrom
|
|
|
| 67 | c4j2nB_ |
|
not modelled |
62.7 |
21 |
PDB header:viral protein
Chain: B: PDB Molecule:gp37;
PDBTitle: crystal structure of mycobacteriophage pukovnik xis
|
|
|
| 68 | c4j2nA_ |
|
not modelled |
62.7 |
21 |
PDB header:viral protein
Chain: A: PDB Molecule:gp37;
PDBTitle: crystal structure of mycobacteriophage pukovnik xis
|
|
|
| 69 | d1w25a3 |
|
not modelled |
62.1 |
11 |
Fold:Ferredoxin-like
Superfamily:Nucleotide cyclase
Family:GGDEF domain |
|
|
| 70 | c4urgB_ |
|
not modelled |
59.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:diguanylate cyclase;
PDBTitle: crystal structure of ggdef domain from t.maritima (active-like dimer)
|
|
|
| 71 | c3tvkA_ |
|
not modelled |
58.3 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:diguanylate cyclase dgcz;
PDBTitle: diguanylate cyclase domain of dgcz
|
|
|
| 72 | d1v6ta_ |
|
not modelled |
57.9 |
16 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 73 | c2x5eA_ |
|
not modelled |
54.8 |
20 |
PDB header:unknown function
Chain: A: PDB Molecule:upf0271 protein pa4511;
PDBTitle: crystal structure of the hypothetical protein pa4511 from2 pseudomonas aeruginosa
|
|
|
| 74 | c3qyyB_ |
|
not modelled |
51.6 |
11 |
PDB header:signaling protein/inhibitor
Chain: B: PDB Molecule:response regulator;
PDBTitle: a novel interaction mode between a microbial ggdef domain and the bis-2 (3, 5 )-cyclic di-gmp
|
|
|
| 75 | c3ungC_ |
|
not modelled |
49.4 |
12 |
PDB header:unknown function
Chain: C: PDB Molecule:cmr2dhd;
PDBTitle: structure of the cmr2 subunit of the crispr rna silencing complex
|
|
|
| 76 | d2dfaa1 |
|
not modelled |
44.7 |
23 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 77 | c3iwfA_ |
|
not modelled |
43.1 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator rpir family;
PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
|
|
|
| 78 | c4dmzB_ |
|
not modelled |
41.6 |
8 |
PDB header:nucleotide-binding protein
Chain: B: PDB Molecule:putative uncharacterized protein peld;
PDBTitle: peld 156-455 from pseudomonas aeruginosa pa14, apo form
|
|
|
| 79 | c4w8yA_ |
|
not modelled |
39.1 |
12 |
PDB header:rna binding protein
Chain: A: PDB Molecule:crispr system cmr subunit cmr2;
PDBTitle: structure of full length cmr2 from pyrococcus furiosus (manganese2 bound form)
|
|
|
| 80 | c5euhA_ |
|
not modelled |
35.9 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:putative ggdef domain membrane protein;
PDBTitle: crystal structure of the c-di-gmp-bound ggdef domain of p. fluorescens2 gcbc
|
|
|
| 81 | c3i5cA_ |
|
not modelled |
35.2 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:fusion of general control protein gcn4 and wspr response
PDBTitle: crystal structure of a fusion protein containing the leucine zipper of2 gcn4 and the ggdef domain of wspr from pseudomonas aeruginosa
|
|
|
| 82 | c4ua2B_ |
|
not modelled |
29.6 |
16 |
PDB header:dna binding protein
Chain: B: PDB Molecule:regulatory protein;
PDBTitle: crystal structure of dual function transcriptional regulator merr from2 bacillus megaterium mb1
|
|
|
| 83 | c3qaoA_ |
|
not modelled |
28.5 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:merr-like transcriptional regulator;
PDBTitle: the crystal structure of the n-terminal domain of a merr-like2 transcriptional regulator from listeria monocytogenes egd-e
|
|
|
| 84 | d2es9a1 |
|
not modelled |
27.9 |
11 |
Fold:YoaC-like
Superfamily:YoaC-like
Family:YoaC-like |
|
|
| 85 | c5m3cB_ |
|
not modelled |
24.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:diguanylate cyclase;
PDBTitle: structure of the hybrid domain (ggdef-eal) of pa0575 from pseudomonas2 aeruginosa pao1 at 2.8 ang. with gtp and ca2+ bound to the active3 site of the ggdef domain
|
|
|
| 86 | c2y0fD_ |
|
not modelled |
22.9 |
9 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
|
|
| 87 | c4ivnB_ |
|
not modelled |
22.2 |
14 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: the vibrio vulnificus nanr protein complexed with mannac-6p
|
|
|
| 88 | c5xgdA_ |
|
not modelled |
21.5 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:uncharacterized protein pa0861;
PDBTitle: crystal structure of the pas-ggdef-eal domain of pa0861 from2 pseudomonas aeruginosa in complex with gtp
|
|
|
| 89 | c3noyA_ |
|
not modelled |
21.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
|
|
| 90 | d2p7vb1 |
|
not modelled |
20.6 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 91 | c2dg6A_ |
|
not modelled |
19.3 |
14 |
PDB header:gene regulation
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of the putative transcriptional regulator sco55502 from streptomyces coelicolor a3(2)
|
|
|
| 92 | c3pjwA_ |
|
not modelled |
16.7 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:cyclic dimeric gmp binding protein;
PDBTitle: structure of pseudomonas fluorescence lapd ggdef-eal dual domain, i23
|
|
|
| 93 | c3i5bA_ |
|
not modelled |
16.0 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:wspr response regulator;
PDBTitle: crystal structure of the isolated ggdef domain of wpsr from2 pseudomonas aeruginosa
|
|
|
| 94 | c3t72o_ |
|
not modelled |
15.8 |
19 |
PDB header:transcription/dna
Chain: O: PDB Molecule:pho box dna (strand 1);
PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
|
|
|
| 95 | c5xqlA_ |
|
not modelled |
15.2 |
6 |
PDB header:transcription
Chain: A: PDB Molecule:multidrug-efflux transporter 1 regulator;
PDBTitle: crystal structure of a pseudomonas aeruginosa transcriptional2 regulator
|
|
|
| 96 | c2o3fC_ |
|
not modelled |
14.0 |
24 |
PDB header:transcription
Chain: C: PDB Molecule:putative hth-type transcriptional regulator ybbh;
PDBTitle: structural genomics, the crystal structure of the n-2 terminal domain of the putative transcriptional regulator3 ybbh from bacillus subtilis subsp. subtilis str. 168.
|
|
|
| 97 | d2o3fa1 |
|
not modelled |
14.0 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:RpiR-like |
|
|
| 98 | d2tssa1 |
|
not modelled |
13.8 |
33 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Superantigen toxins, N-terminal domain |
|
|
| 99 | c2cwbA_ |
|
not modelled |
13.3 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:chimera of immunoglobulin g binding protein g
PDBTitle: solution structure of the ubiquitin-associated domain of2 human bmsc-ubp and its complex with ubiquitin
|
|
|