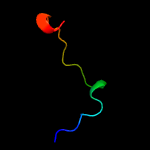
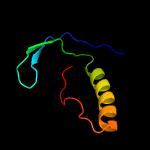
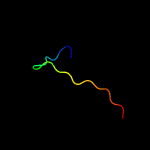
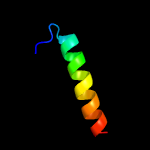
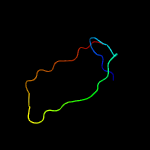
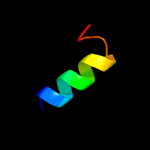
1 c6amgA_
23.5
26
PDB header: metal binding proteinChain: A: PDB Molecule: cytochrome p460;PDBTitle: cyt p460 of nitrosomonas sp. al212
2 c3sc0A_
18.9
30
PDB header: oxidoreductaseChain: A: PDB Molecule: methylmalonic aciduria and homocystinuria type c protein;PDBTitle: crystal structure of mmachc (1-238), a human b12 processing enzyme,2 complexed with methylcobalamin
3 c1kn7A_
11.8
78
PDB header: membrane proteinChain: A: PDB Molecule: voltage-gated potassium channel protein kv1.4;PDBTitle: solution structure of the tandem inactivation domain2 (residues 1-75) of potassium channel rck4 (kv1.4)
4 c4blgB_
11.6
45
PDB header: viral proteinChain: B: PDB Molecule: latency-associated nuclear antigen;PDBTitle: crystal structure of mhv-68 latency-associated nuclear antigen (lana)2 c-terminal dna binding domain
5 c4k2jB_
11.1
64
PDB header: dna binding protein, viral proteinChain: B: PDB Molecule: kshv (hhv-8) latency-associated nuclear antigen (lana);PDBTitle: decameric ring structure of kshv (hhv-8) latency-associated nuclear2 antigen (lana) dna binding domain
6 c3cvoA_
10.7
31
PDB header: transferaseChain: A: PDB Molecule: methyltransferase-like protein of unknown function;PDBTitle: crystal structure of a methyltransferase-like protein (spo2022) from2 silicibacter pomeroyi dss-3 at 1.80 a resolution
7 c6db1A_
10.3
33
PDB header: transferaseChain: A: PDB Molecule: putative methyl-accepting chemotaxis protein;PDBTitle: 2.0 angstrom resolution crystal structure of n-terminal ligand-binding2 domain of putative methyl-accepting chemotaxis protein from3 salmonella enterica
8 c4wl2F_
10.1
33
PDB header: hydrolaseChain: F: PDB Molecule: putative exported choloylglycine hydrolase;PDBTitle: structure of penicillin v acylase from pectobacterium atrosepticum
9 c2rkqA_
10.0
19
PDB header: immune systemChain: A: PDB Molecule: peptidoglycan-recognition protein-sd;PDBTitle: crystal structure of drosophila peptidoglycan recognition2 protein sd (pgrp-sd)
10 c2yuyA_
9.4
21
PDB header: signaling proteinChain: A: PDB Molecule: rho gtpase activating protein 21;PDBTitle: solution structure of pdz domain of rho gtpase activating2 protein 21
11 c6hiuA_
9.1
26
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p460;PDBTitle: cytochrome p460 from methylococcus capsulatus (bath)
12 c3oc3B_
7.7
54
PDB header: hydrolase/transcriptionChain: B: PDB Molecule: helicase mot1;PDBTitle: crystal structure of the mot1 n-terminal domain in complex with tbp
13 d16vpa_
7.6
55
Fold: Conserved core of transcriptional regulatory protein vp16Superfamily: Conserved core of transcriptional regulatory protein vp16Family: Conserved core of transcriptional regulatory protein vp16
14 c4h7wA_
7.4
19
PDB header: unknown functionChain: A: PDB Molecule: upf0406 protein c16orf57;PDBTitle: crystal structure of human c16orf57
15 c6gw6A_
7.3
60
PDB header: toxinChain: A: PDB Molecule: res toxin;PDBTitle: structure of the pseudomonas putida res-xre toxin-antitoxin complex
16 d2bopa_
7.1
33
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
17 d2ia9a1
6.9
20
Fold: SpoVG-likeSuperfamily: SpoVG-likeFamily: SpoVG-like
18 c3somO_
6.7
35
PDB header: oxidoreductaseChain: O: PDB Molecule: methylmalonic aciduria and homocystinuria type c protein;PDBTitle: crystal structure of human mmachc
19 c1i26A_
6.5
44
PDB header: toxinChain: A: PDB Molecule: ptu-1;PDBTitle: solution structure of ptu-1, a toxin from the assassin bugs2 peirates turpis that blocks the voltage sensitive calcium3 channel n-type
20 d1gpqa_
6.4
50
Fold: Inhibitor of vertebrate lysozyme, IvySuperfamily: Inhibitor of vertebrate lysozyme, IvyFamily: Inhibitor of vertebrate lysozyme, Ivy
21 d1nixa_
not modelled
6.3
55
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
22 c6gftA_
not modelled
6.0
55
PDB header: toxinChain: A: PDB Molecule: cyriotoxin-1a;PDBTitle: antinociceptive evaluation of cyriotoxin-1a, the first toxin purified2 from cyriopagopus schioedtei spider venom
23 c4je3B_
not modelled
5.9
50
PDB header: cell cycleChain: B: PDB Molecule: central kinetochore subunit chl4;PDBTitle: an iml3-chl4 heterodimer links the core centromere to factors required2 for accurate chromosome segregation
24 c4xa6C_
not modelled
5.7
70
PDB header: motor proteinChain: C: PDB Molecule: gp7-myh7(1777-1855)-eb1 chimera protein;PDBTitle: crystal structure of the coiled-coil surrounding skip 4 of myh7
25 c4xa6B_
not modelled
5.6
70
PDB header: motor proteinChain: B: PDB Molecule: gp7-myh7(1777-1855)-eb1 chimera protein;PDBTitle: crystal structure of the coiled-coil surrounding skip 4 of myh7
26 c5j9hA_
not modelled
5.4
36
PDB header: viral proteinChain: A: PDB Molecule: envelopment polyprotein;PDBTitle: crystal structure of glycoprotein c from puumala virus in the post-2 fusion conformation (ph 8.0)
27 c6fcxA_
not modelled
5.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: methylenetetrahydrofolate reductase;PDBTitle: structure of human 5,10-methylenetetrahydrofolate reductase (mthfr)
28 c3nctC_
not modelled
5.4
25
PDB header: dna binding protein, chaperoneChain: C: PDB Molecule: protein psib;PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
29 c5zk4D_
not modelled
5.1
24
PDB header: transferaseChain: D: PDB Molecule: disa protein;PDBTitle: the structure of dszs acyltransferase with carrier protein
30 c5m9eA_
not modelled
5.1
55
PDB header: cell cycleChain: A: PDB Molecule: microtubule integrity protein mal3;PDBTitle: interactions between the mal3 eb1-like domain and dis1
31 c2n3pA_
not modelled
5.1
63
PDB header: toxinChain: A: PDB Molecule: asteropsin_g;PDBTitle: solution nmr structure of asteropsin g from marine sponge asteropus