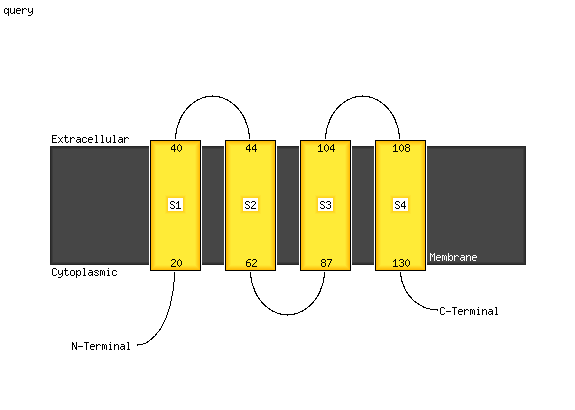
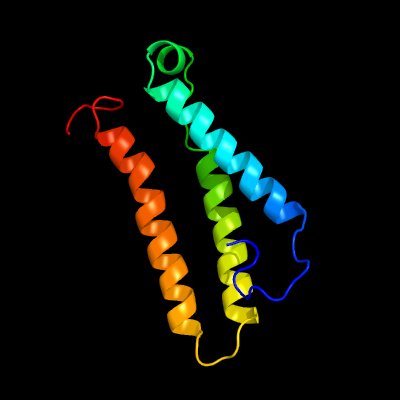
1 c3ug9A_
62.7
17
PDB header: membrane proteinChain: A: PDB Molecule: archaeal-type opsin 1, archaeal-type opsin 2;PDBTitle: crystal structure of the closed state of channelrhodopsin
2 c4ezdB_
61.9
21
PDB header: transport proteinChain: B: PDB Molecule: urea transporter 1;PDBTitle: crystal structure of the ut-b urea transporter from bos taurus bound2 to selenourea
3 c5zihA_
57.7
11
PDB header: membrane proteinChain: A: PDB Molecule: sensory opsin a,chrimson;PDBTitle: crystal structure of the red light-activated channelrhodopsin2 chrimson.
4 c3k3gA_
57.3
19
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
5 c6eyuA_
20.1
13
PDB header: membrane proteinChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: crystal structure of the inward h(+) pump xenorhodopsin
6 d1fp1d1
17.6
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain
7 c6csmC_
16.9
10
PDB header: membrane proteinChain: C: PDB Molecule: gtacr1;PDBTitle: crystal structure of the natural light-gated anion channel gtacr1
8 c5tr1A_
16.2
19
PDB header: transport proteinChain: A: PDB Molecule: chloride channel protein;PDBTitle: cryo-electron microscopy structure of a bovine clc-k chloride channel,2 alternate (class 2) conformation
9 c4mt1A_
15.0
22
PDB header: membrane protein, tranport proteinChain: A: PDB Molecule: drug efflux protein;PDBTitle: crystal structure of the neisseria gonorrhoeae mtrd inner membrane2 multidrug efflux pump
10 c2l35A_
11.5
28
PDB header: protein bindingChain: A: PDB Molecule: dap12-nkg2c_tm;PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
11 c5iwsA_
11.3
13
PDB header: transferaseChain: A: PDB Molecule: protein-n(pi)-phosphohistidine-sugar phosphotransferasePDBTitle: crystal structure of the transporter malt, the eiic domain from the2 maltose-specific phosphotransferase system
12 c5vaqC_
10.8
78
PDB header: signaling proteinChain: C: PDB Molecule: fibroblast growth factor 21;PDBTitle: crystal structure of beta-klotho in complex with fgf21ct
13 c1o7dD_
10.3
75
PDB header: hydrolaseChain: D: PDB Molecule: lysosomal alpha-mannosidase;PDBTitle: the structure of the bovine lysosomal a-mannosidase suggests a novel2 mechanism for low ph activation
14 c6qvcB_
9.0
19
PDB header: membrane proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: cryoem structure of the human clc-1 chloride channel, cbs state 1
15 c3orgB_
8.9
22
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
16 c6o7ua_
8.7
26
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
17 d1kyza1
8.1
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain
18 c4jspA_
8.1
14
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase mtor;PDBTitle: structure of mtordeltan-mlst8-atpgammas-mg complex
19 c2n90B_
6.5
26
PDB header: transferaseChain: B: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
20 c2n90A_
6.5
26
PDB header: transferaseChain: A: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
21 c6coyB_
not modelled
6.3
19
PDB header: transport proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: human clc-1 chloride ion channel, transmembrane domain
22 c2jwaA_
not modelled
6.2
67
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
23 c2ks1A_
not modelled
6.2
67
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
24 c5o60b_
not modelled
5.8
21
PDB header: ribosomeChain: B: PDB Molecule: 5s rrna;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
25 c5ldwc_
not modelled
5.5
43
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class1
26 c5lc5c_
not modelled
5.5
43
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class2
27 c2mltB_
not modelled
5.4
53
PDB header: toxin (hemolytic polypeptide)Chain: B: PDB Molecule: melittin;PDBTitle: melittin
28 c2mltA_
not modelled
5.4
53
PDB header: toxin (hemolytic polypeptide)Chain: A: PDB Molecule: melittin;PDBTitle: melittin
29 c6o4mB_
not modelled
5.4
53
PDB header: toxinChain: B: PDB Molecule: melittin;PDBTitle: racemic melittin
30 c6dstA_
not modelled
5.4
53
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: recombinant melittin
31 c6o4mA_
not modelled
5.4
53
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: racemic melittin
32 c6d0nA_
not modelled
5.2
23
PDB header: transport proteinChain: A: PDB Molecule: clc-type fluoride/proton antiporter;PDBTitle: crystal structure of a clc-type fluoride/proton antiporter, v319g2 mutant