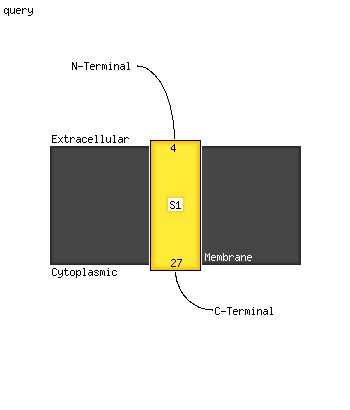
1 c6fkfd_
100.0
18
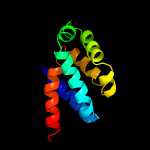
PDB header: membrane proteinChain: D: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 1
2 c6rdlP_
100.0
23
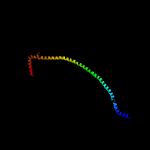
PDB header: proton transportChain: P: PDB Molecule: mitochondrial atp synthase subunit oscp;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1b,2 monomer-masked refinement
3 c5t4oL_
100.0
19
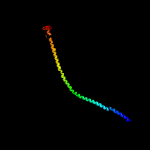
PDB header: hydrolaseChain: L: PDB Molecule: atp synthase subunit delta;PDBTitle: autoinhibited e. coli atp synthase state 1
4 c6fkib_
100.0
11
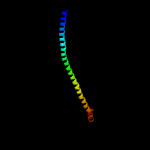
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
5 c5t4oJ_
99.9
26
PDB header: hydrolaseChain: J: PDB Molecule: atp synthase subunit b;PDBTitle: autoinhibited e. coli atp synthase state 1
6 c6fkip_
99.9
11
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
7 c6b8ho_
99.9
21
PDB header: membrane proteinChain: O: PDB Molecule: atp synthase subunit 5, mitochondrial;PDBTitle: mosaic model of yeast mitochondrial atp synthase monomer
8 c2wssS_
99.9
22
PDB header: hydrolaseChain: S: PDB Molecule: atp synthase subunit o, mitochondrial;PDBTitle: the structure of the membrane extrinsic region of bovine atp synthase
9 c5lqzU_
99.9
17
PDB header: hydrolaseChain: U: PDB Molecule: atp synthase oscp subunit;PDBTitle: structure of f-atpase from pichia angusta, state1
10 c6b8hb_
99.9
13
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit alpha, mitochondrial;PDBTitle: mosaic model of yeast mitochondrial atp synthase monomer
11 c5dn6H_
99.8
17
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase subunit delta;PDBTitle: atp synthase from paracoccus denitrificans
12 c2jmxA_
99.7
23
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase o subunit, mitochondrial;PDBTitle: oscp-nt (1-120) in complex with n-terminal (1-25) alpha2 subunit from f1-atpase
13 c6rdvP_
99.7
22
PDB header: proton transportChain: P: PDB Molecule: mitochondrial atp synthase subunit oscp;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 focussed refinement of f1 head and rotor
14 d1abva_
99.7
13
Fold: ATPD N-terminal domain-likeSuperfamily: N-terminal domain of the delta subunit of the F1F0-ATP synthaseFamily: N-terminal domain of the delta subunit of the F1F0-ATP synthase
15 c2a7uB_
99.7
13
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase delta chain;PDBTitle: nmr solution structure of the e.coli f-atpase delta subunit n-terminal2 domain in complex with alpha subunit n-terminal 22 residues
16 c6j5ib_
99.5
9
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit alpha, mitochondrial;PDBTitle: cryo-em structure of the mammalian dp-state atp synthase
17 c5lqyV_
98.5
18
PDB header: hydrolaseChain: V: PDB Molecule: atp synthase subunit b;PDBTitle: structure of f-atpase from pichia angusta, in state2
18 c5arhT_
98.5
11
PDB header: hydrolaseChain: T: PDB Molecule: atp synthase f(0) complex subunit b1, mitochondrial;PDBTitle: bovine mitochondrial atp synthase state 2a
19 d1l2pa_
98.4
27
Fold: Single transmembrane helixSuperfamily: F1F0 ATP synthase subunit B, membrane domainFamily: F1F0 ATP synthase subunit B, membrane domain
20 c4dt0A_
97.6
20
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit e;PDBTitle: the structure of the peripheral stalk subunit e from pyrococcus2 horikoshii
21 c3k5bE_
not modelled
97.5
19
PDB header: hydrolaseChain: E: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
22 c1b9uA_
not modelled
97.0
21
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
23 c4dl0J_
not modelled
97.0
7
PDB header: hydrolaseChain: J: PDB Molecule: v-type proton atpase subunit e;PDBTitle: crystal structure of the heterotrimeric egchead peripheral stalk2 complex of the yeast vacuolar atpase
24 c2khkA_
not modelled
95.1
34
PDB header: transport proteinChain: A: PDB Molecule: atp synthase subunit b;PDBTitle: nmr solution structure of the b30-82 domain of subunit b of2 escherichia coli f1fo atp synthase
25 c5b0oG_
not modelled
94.9
8
PDB header: hydrolase/motor proteinChain: G: PDB Molecule: flagellar assembly protein flih;PDBTitle: structure of the flih-flii complex
26 d2dm9a1
not modelled
93.6
15
Fold: FwdE/GAPDH domain-likeSuperfamily: V-type ATPase subunit E-likeFamily: V-type ATPase subunit E
27 c2dm9B_
not modelled
93.6
15
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of ph1978 from pyrococcus horikoshii ot3
28 c5b0oH_
not modelled
92.7
10
PDB header: hydrolase/motor proteinChain: H: PDB Molecule: flagellar assembly protein flih;PDBTitle: structure of the flih-flii complex
29 c2oarA_
not modelled
92.4
13
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
30 c3uz0A_
not modelled
91.4
16
PDB header: transport proteinChain: A: PDB Molecule: stage iii sporulation protein ah;PDBTitle: crystal structure of spoiiiah and spoiiq complex
31 c6b2zb_
not modelled
91.2
20
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit c, mitochondrial;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
32 c3vkgA_
not modelled
90.6
16
PDB header: motor proteinChain: A: PDB Molecule: dynein heavy chain, cytoplasmic;PDBTitle: x-ray structure of an mtbd truncation mutant of dynein motor domain
33 c3lg8B_
not modelled
90.2
13
PDB header: hydrolaseChain: B: PDB Molecule: a-type atp synthase subunit e;PDBTitle: crystal structure of the c-terminal part of subunit e (e101-206) from2 methanocaldococcus jannaschii of a1ao atp synthase
34 d2oara1
not modelled
89.9
16
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel
35 c1y4cA_
not modelled
88.6
18
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
36 c2k88A_
not modelled
88.0
28
PDB header: hydrolaseChain: A: PDB Molecule: vacuolar proton pump subunit g;PDBTitle: association of subunit d (vma6p) and e (vma4p) with g2 (vma10p) and the nmr solution structure of subunit g (g1-3 59) of the saccharomyces cerevisiae v1vo atpase
37 c3tufA_
not modelled
87.5
16
PDB header: signaling proteinChain: A: PDB Molecule: stage iii sporulation protein ah;PDBTitle: structure of the spoiiq-spoiiiah pore forming complex.
38 c2zv4O_
not modelled
86.4
10
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
39 c4y7jE_
not modelled
84.8
30
PDB header: membrane protein,transport proteinChain: E: PDB Molecule: large conductance mechanosensitive channel protein,PDBTitle: structure of an archaeal mechanosensitive channel in expanded state
40 c3k5bB_
not modelled
84.1
15
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase, subunit (vapc-therm);PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
41 c4dl0G_
not modelled
83.3
19
PDB header: hydrolaseChain: G: PDB Molecule: v-type proton atpase subunit g;PDBTitle: crystal structure of the heterotrimeric egchead peripheral stalk2 complex of the yeast vacuolar atpase
42 c6flnE_
not modelled
83.3
13
PDB header: protein bindingChain: E: PDB Molecule: e3 ubiquitin/isg15 ligase trim25;PDBTitle: crystal structure of the human trim25 coiled-coil and pryspry domains
43 c3hzqA_
not modelled
82.9
22
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: structure of a tetrameric mscl in an expanded intermediate state
44 c4cg4D_
not modelled
71.9
7
PDB header: actin-binding proteinChain: D: PDB Molecule: pyrin;PDBTitle: crystal structure of the chs-b30.2 domains of trim20
45 c3wolB_
not modelled
65.5
9
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidyl aminopeptidase bii;PDBTitle: crystal structure of the dap bii dipeptide complex i
46 c2kk7A_
not modelled
63.8
34
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit e;PDBTitle: nmr solution structure of the n terminal domain of subunit e2 (e1-52) of a1ao atp synthase from methanocaldococcus3 jannaschii
47 c2rddB_
not modelled
63.6
6
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
48 c5jxfA_
not modelled
63.5
12
PDB header: hydrolaseChain: A: PDB Molecule: asp/glu-specific dipeptidyl-peptidase;PDBTitle: crystal structure of flavobacterium psychrophilum dpp11 in complex2 with dipeptide arg-asp
49 c2ayaA_
not modelled
62.0
25
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii subunit tau;PDBTitle: solution structure of the c-terminal 14 kda domain of the2 tau subunit from escherichia coli dna polymerase iii
50 c4nqjB_
not modelled
57.3
8
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase trim69;PDBTitle: structure of coiled-coil domain
51 c3ojaB_
not modelled
57.2
11
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
52 c6qajB_
not modelled
51.7
12
PDB header: nuclear proteinChain: B: PDB Molecule: endolysin,transcription intermediary factor 1-beta;PDBTitle: structure of the tripartite motif of kap1/trim28
53 c4xtbA_
not modelled
44.9
23
PDB header: transport proteinChain: A: PDB Molecule: calcium uniporter protein, mitochondrial;PDBTitle: crystal structure of the n-terminal domain of the human mitochondrial2 calcium uniporter
54 c1qcrD_
not modelled
42.4
12
PDB header: oxidoreductaseChain: D: PDB Molecule: ubiquinol cytochrome c oxidoreductase;PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
55 c3ipzA_
not modelled
40.9
16
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-s14, chloroplastic;PDBTitle: crystal structure of arabidopsis monothiol glutaredoxin atgrxcp
56 c3vkhA_
not modelled
39.4
12
PDB header: motor proteinChain: A: PDB Molecule: dynein heavy chain, cytoplasmic;PDBTitle: x-ray structure of a functional full-length dynein motor domain
57 d2ou3a1
not modelled
39.0
14
Fold: TerB-likeSuperfamily: TerB-likeFamily: COG3793-like
58 c2lseA_
not modelled
32.4
13
PDB header: de novo proteinChain: A: PDB Molecule: four helix bundle protein;PDBTitle: solution nmr structure of de novo designed four helix bundle protein,2 northeast structural genomics consortium (nesg) target or188
59 c4kqtA_
not modelled
30.6
11
PDB header: chaperoneChain: A: PDB Molecule: putative outer membrane chaperone (omph-like);PDBTitle: crystal structure of a putative outer membrane chaperone (omph-like)2 (cc_1914) from caulobacter crescentus cb15 at 2.83 a resolution (psi3 community target, shapiro)
60 c2k6iA_
not modelled
30.3
31
PDB header: structural proteinChain: A: PDB Molecule: uncharacterized protein mj0223;PDBTitle: the domain features of the peripheral stalk subunit h of the2 methanogenic a1ao atp synthase and the nmr solution3 structure of h1-47
61 c4b2qt_
not modelled
30.1
10
PDB header: hydrolaseChain: T: PDB Molecule: atp synthase subunit b, mitochondrial;PDBTitle: model of the yeast f1fo-atp synthase dimer based on subtomogram2 average
62 c1u2mC_
not modelled
29.0
8
PDB header: chaperoneChain: C: PDB Molecule: histone-like protein hlp-1;PDBTitle: crystal structure of skp
63 c6b2zL_
not modelled
28.1
21
PDB header: membrane proteinChain: L: PDB Molecule: atp synthase protein 8;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
64 c6b2zA_
not modelled
28.1
21
PDB header: membrane proteinChain: A: PDB Molecule: atp synthase protein 8;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
65 d2al1a2
not modelled
27.8
42
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
66 d2uubc1
not modelled
27.2
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)
67 c1ei3E_
not modelled
26.4
11
PDB header: blood clottingChain: E: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
68 d1sqwa2
not modelled
24.8
13
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: Nip7p homolog, N-terminal domain
69 d2np5a2
not modelled
24.2
15
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
70 c3rfrI_
not modelled
24.1
12
PDB header: oxidoreductaseChain: I: PDB Molecule: pmob;PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
71 c3ghgI_
not modelled
24.0
5
PDB header: blood clottingChain: I: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of human fibrinogen
72 c5zazA_
not modelled
23.9
15
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: solution structure of integrin b2 monomer tranmembrane domain in2 bicelle
73 c3jacA_
not modelled
22.9
20
PDB header: metal transportChain: A: PDB Molecule: piezo-type mechanosensitive ion channel component 1;PDBTitle: cryo-em study of a channel
74 c4k08A_
not modelled
22.7
17
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis sensory transducer;PDBTitle: periplasmic sensor domain of chemotaxis protein, adeh_3718
75 c3lvgD_
not modelled
22.4
11
PDB header: structural proteinChain: D: PDB Molecule: clathrin light chain b;PDBTitle: crystal structure of a clathrin heavy chain and clathrin light chain2 complex
76 c3cjeA_
not modelled
21.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: osmc-like protein;PDBTitle: crystal structure of an osmc-like hydroperoxide resistance protein2 (jann_2040) from jannaschia sp. ccs1 at 1.70 a resolution
77 c4pd3B_
not modelled
21.7
8
PDB header: contractile proteinChain: B: PDB Molecule: nonmuscle myosin heavy chain b, alpha-actinin a chimeraPDBTitle: crystal structure of rigor-like human nonmuscle myosin-2b
78 c4qs9A_
not modelled
21.2
12
PDB header: transferaseChain: A: PDB Molecule: hexokinase-1;PDBTitle: arabidopsis hexokinase 1 (athxk1) mutant s177a structure in glucose-2 bound form
79 c3gx8A_
not modelled
20.8
25
PDB header: electron transportChain: A: PDB Molecule: monothiol glutaredoxin-5, mitochondrial;PDBTitle: structural and biochemical characterization of yeast2 monothiol glutaredoxin grx5
80 c1deqF_
not modelled
20.7
10
PDB header: blood clottingChain: F: PDB Molecule: fibrinogen (gamma chain);PDBTitle: the crystal structure of modified bovine fibrinogen (at ~42 angstrom resolution)
81 c3vkhB_
not modelled
20.1
13
PDB header: motor proteinChain: B: PDB Molecule: dynein heavy chain, cytoplasmic;PDBTitle: x-ray structure of a functional full-length dynein motor domain
82 c5mlc9_
not modelled
19.3
13
PDB header: ribosomeChain: 9: PDB Molecule: ribosome-recycling factor, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
83 c5xauD_
not modelled
18.5
18
PDB header: cell adhesionChain: D: PDB Molecule: laminin subunit alpha-5;PDBTitle: crystal structure of integrin binding fragment of laminin-511
84 d1bg3a2
not modelled
18.2
13
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
85 c3jacC_
not modelled
18.2
20
PDB header: metal transportChain: C: PDB Molecule: piezo-type mechanosensitive ion channel component 1;PDBTitle: cryo-em study of a channel
86 c2qzvB_
not modelled
17.5
10
PDB header: structural proteinChain: B: PDB Molecule: major vault protein;PDBTitle: draft crystal structure of the vault shell at 9 angstroms2 resolution
87 c2jacA_
not modelled
17.3
10
PDB header: electron transportChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: glutaredoxin grx1p c30s mutant from yeast
88 c3spuB_
not modelled
16.8
22
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase ndm-1;PDBTitle: apo ndm-1 crystal structure
89 c3ghgK_
not modelled
16.7
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
90 c3n5iC_
not modelled
16.6
11
PDB header: hydrolaseChain: C: PDB Molecule: beta-peptidyl aminopeptidase;PDBTitle: crystal structure of the precursor (s250a mutant) of the n-terminal2 beta-aminopeptidase bapa
91 c5ew5C_
not modelled
16.5
9
PDB header: hydrolaseChain: C: PDB Molecule: colicin-e9;PDBTitle: crystal structure of colicin e9 in complex with its immunity protein2 im9
92 c3c1sA_
not modelled
15.9
10
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structure of grx1 in glutathionylated form
93 c2kz9A_
not modelled
15.8
8
PDB header: proton transportChain: A: PDB Molecule: v-type proton atpase subunit e;PDBTitle: structure of e1-69 of yeast v-atpase
94 d1ts9a_
not modelled
15.5
24
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
95 c2rmzA_
not modelled
15.4
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
96 d2akza2
not modelled
15.4
23
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
97 c5nugB_
not modelled
15.2
14
PDB header: motor proteinChain: B: PDB Molecule: cytoplasmic dynein 1 heavy chain 1;PDBTitle: motor domains from human cytoplasmic dynein-1 in the phi-particle2 conformation
98 c5t77A_
not modelled
15.1
17
PDB header: transport proteinChain: A: PDB Molecule: putative lipid ii flippase murj;PDBTitle: crystal structure of the mop flippase murj
99 c2k1aA_
not modelled
15.1
20
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment