| 1 | c6fkig_
|
|
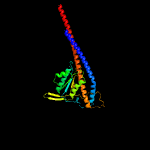 |
100.0 |
36 |
PDB header:membrane protein
Chain: G: PDB Molecule:atp synthase subunit c, chloroplastic;
PDBTitle: chloroplast f1fo conformation 3
|
|
|
|
| 2 | c3oaaO_
|
|
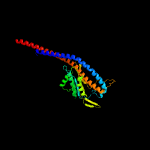 |
100.0 |
35 |
PDB header:hydrolase/transport protein
Chain: O: PDB Molecule:atp synthase gamma chain;
PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
|
|
|
|
| 3 | c6q45G_
|
|
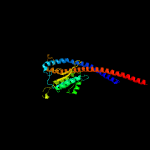 |
100.0 |
35 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: f1-atpase from fusobacterium nucleatum
|
|
|
|
| 4 | c5dn6G_
|
|
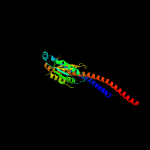 |
100.0 |
37 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: atp synthase from paracoccus denitrificans
|
|
|
|
| 5 | c6re1S_
|
|
 |
100.0 |
27 |
PDB header:proton transport
Chain: S: PDB Molecule:atp synthase gamma chain, mitochondrial;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 2a,2 focussed refinement of f1 head and rotor
|
|
|
|
| 6 | c2w6hG_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase during2 controlled dehydration: hydration state 4a.
|
|
|
|
| 7 | c5lqxG_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma subunit;
PDBTitle: structure of f-atpase from pichia angusta, state3
|
|
|
|
| 8 | c6f5dG_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma subunit;
PDBTitle: trypanosoma brucei f1-atpase
|
|
|
|
| 9 | c2xokG_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a resolution
|
|
|
|
| 10 | c4xd7G_
|
|
 |
100.0 |
41 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: structure of thermophilic f1-atpase inhibited by epsilon subunit
|
|
|
|
| 11 | c5zwlG_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: crystal structure of the gamma - epsilon complex of photosynthetic2 cyanobacterial f1-atpase
|
|
|
|
| 12 | c6focG_
|
|
 |
100.0 |
88 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: f1-atpase from mycobacterium smegmatis
|
|
|
|
| 13 | d2jdig1
|
|
 |
100.0 |
43 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
|
|
|
| 14 | d1fs0g_
|
|
 |
100.0 |
33 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
|
|
|
| 15 | c2qe7G_
|
|
 |
100.0 |
39 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
|
|
|
|
| 16 | c2w6jG_
|
|
 |
100.0 |
45 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase during2 controlled dehydration: hydration state 5.
|
|
|
|
| 17 | c3fksY_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: Y: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: yeast f1 atpase in the absence of bound nucleotides
|
|
|
|
| 18 | d1mabg_
|
|
 |
100.0 |
44 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
|
|
|
| 19 | c3re1B_
|
|
 |
68.0 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthetase;
PDBTitle: crystal structure of uroporphyrinogen iii synthase from pseudomonas2 syringae pv. tomato dc3000
|
|
|
|
| 20 | c4es6A_
|
|
 |
52.3 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of hemd (pa5259) from pseudomonas aeruginosa (pao1)2 at 2.22 a resolution
|
|
|
|
| 21 | c3k8vB_ |
|
not modelled |
50.8 |
19 |
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
|
|
|
| 22 | d1wd7a_ |
|
not modelled |
49.3 |
14 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
|
|
| 23 | c3gitA_ |
|
not modelled |
45.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase/acetyl-coa synthase subunit
PDBTitle: crystal structure of a truncated acetyl-coa synthase
|
|
|
| 24 | d1ru3a_ |
|
not modelled |
44.8 |
23 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Acetyl-CoA synthase |
|
|
| 25 | d1oaoc_ |
|
not modelled |
41.2 |
21 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Acetyl-CoA synthase |
|
|
| 26 | c3d8tB_ |
|
not modelled |
35.1 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
|
|
| 27 | c5vayF_ |
|
not modelled |
33.1 |
28 |
PDB header:apoptosis
Chain: F: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 t108d bh3 domain
|
|
|
| 28 | c5vayE_ |
|
not modelled |
33.1 |
28 |
PDB header:apoptosis
Chain: E: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 t108d bh3 domain
|
|
|
| 29 | c5vaxE_ |
|
not modelled |
33.1 |
28 |
PDB header:apoptosis
Chain: E: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 30 | c6dcoD_ |
|
not modelled |
30.3 |
28 |
PDB header:apoptosis
Chain: D: PDB Molecule:beclin-1;
PDBTitle: bcl-xl complex with beclin 1 bh3 domain t108d
|
|
|
| 31 | c5vb4D_ |
|
not modelled |
30.1 |
28 |
PDB header:apoptosis
Chain: D: PDB Molecule:beclin-1;
PDBTitle: bcl-xl complex with beclin 1 bh3 domain t108d
|
|
|
| 32 | c5vaxG_ |
|
not modelled |
30.0 |
28 |
PDB header:apoptosis
Chain: G: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 33 | c5vauF_ |
|
not modelled |
28.7 |
28 |
PDB header:apoptosis
Chain: F: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 34 | d1qw2a_ |
|
not modelled |
27.9 |
23 |
Fold:Hypothetical protein Ta1206
Superfamily:Hypothetical protein Ta1206
Family:Hypothetical protein Ta1206 |
|
|
| 35 | c6hr5A_ |
|
not modelled |
27.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-l-rhamnosidase/sulfatase (gh78);
PDBTitle: structure of the s1_25 family sulfatase module of the rhamnosidase2 fa22250 from formosa agariphila
|
|
|
| 36 | c2p1lB_ |
|
not modelled |
26.3 |
28 |
PDB header:apoptosis
Chain: B: PDB Molecule:beclin 1;
PDBTitle: structure of the bcl-xl:beclin 1 complex
|
|
|
| 37 | c2p1lF_ |
|
not modelled |
26.3 |
28 |
PDB header:apoptosis
Chain: F: PDB Molecule:beclin 1;
PDBTitle: structure of the bcl-xl:beclin 1 complex
|
|
|
| 38 | c2p1lD_ |
|
not modelled |
26.3 |
28 |
PDB header:apoptosis
Chain: D: PDB Molecule:beclin 1;
PDBTitle: structure of the bcl-xl:beclin 1 complex
|
|
|
| 39 | c2p1lH_ |
|
not modelled |
26.3 |
28 |
PDB header:apoptosis
Chain: H: PDB Molecule:beclin 1;
PDBTitle: structure of the bcl-xl:beclin 1 complex
|
|
|
| 40 | c5vauE_ |
|
not modelled |
25.0 |
28 |
PDB header:apoptosis
Chain: E: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 41 | c5vaxF_ |
|
not modelled |
25.0 |
28 |
PDB header:apoptosis
Chain: F: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 42 | c5vaxH_ |
|
not modelled |
25.0 |
28 |
PDB header:apoptosis
Chain: H: PDB Molecule:beclin-1;
PDBTitle: bcl-2 complex with beclin 1 bh3 domain
|
|
|
| 43 | c3v47D_ |
|
not modelled |
24.0 |
23 |
PDB header:immune system
Chain: D: PDB Molecule:flagellin;
PDBTitle: crystal structure of the n-terminal fragment of zebrafish tlr5 in2 complex with salmonella flagellin
|
|
|
| 44 | d1sfea2 |
|
not modelled |
23.7 |
50 |
Fold:Ribonuclease H-like motif
Superfamily:Methylated DNA-protein cysteine methyltransferase domain
Family:Methylated DNA-protein cysteine methyltransferase domain |
|
|
| 45 | c2zktB_ |
|
not modelled |
22.2 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: structure of ph0037 protein from pyrococcus horikoshii
|
|
|
| 46 | c3dvuD_ |
|
not modelled |
21.8 |
28 |
PDB header:viral protein/apoptosis
Chain: D: PDB Molecule:beclin-1;
PDBTitle: crystal structure of the complex of murine gamma-herpesvirus 68 bcl-22 homolog m11 and the beclin 1 bh3 domain
|
|
|
| 47 | c5gz5A_ |
|
not modelled |
21.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:snake venom phosphodiesterase (pde);
PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra) in complex with amp
|
|
|
| 48 | c3m8yC_ |
|
not modelled |
21.3 |
23 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphopentomutase;
PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
|
|
|
| 49 | c3lzcA_ |
|
not modelled |
21.1 |
23 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:dph2;
PDBTitle: crystal structure of dph2 from pyrococcus horikoshii
|
|
|
| 50 | d2fefa1 |
|
not modelled |
20.0 |
42 |
Fold:Bromodomain-like
Superfamily:PA2201 N-terminal domain-like
Family:PA2201 N-terminal domain-like |
|
|
| 51 | c3dvuC_ |
|
not modelled |
19.8 |
28 |
PDB header:viral protein/apoptosis
Chain: C: PDB Molecule:beclin-1;
PDBTitle: crystal structure of the complex of murine gamma-herpesvirus 68 bcl-22 homolog m11 and the beclin 1 bh3 domain
|
|
|
| 52 | d2pk8a1 |
|
not modelled |
18.9 |
31 |
Fold:Hypothetical protein PF0899
Superfamily:Hypothetical protein PF0899
Family:Hypothetical protein PF0899 |
|
|
| 53 | c5gz4A_ |
|
not modelled |
18.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:snake venom phosphodiesterase (pde);
PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra)
|
|
|
| 54 | c5i5fA_ |
|
not modelled |
18.8 |
12 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein yejm;
PDBTitle: salmonella global domain 191
|
|
|
| 55 | c2kilA_ |
|
not modelled |
18.6 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: nmr structure of the h103g mutant so2144 h-nox domain from2 shewanella oneidensis in the fe(ii)co ligation state
|
|
|
| 56 | c5xynC_ |
|
not modelled |
17.9 |
17 |
PDB header:dna binding protein
Chain: C: PDB Molecule:suppressor of hu sensitivity involved in recombination
PDBTitle: the crystal structure of csm2-psy3-shu1-shu2 complex from budding2 yeast
|
|
|
| 57 | d1o98a2 |
|
not modelled |
17.8 |
13 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
|
|
| 58 | c2qzuA_ |
|
not modelled |
17.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
|
|
|
| 59 | c4b56A_ |
|
not modelled |
16.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: structure of ectonucleotide pyrophosphatase-phosphodiesterase-12 (npp1)
|
|
|
| 60 | c1pqrA_ |
|
not modelled |
16.4 |
71 |
PDB header:toxin
Chain: A: PDB Molecule:alpha-a-conotoxin eiva;
PDBTitle: solution conformation of alphaa-conotoxin eiva
|
|
|
| 61 | d1auka_ |
|
not modelled |
15.9 |
15 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 62 | d1q77a_ |
|
not modelled |
15.8 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
|
|
| 63 | c3opyB_ |
|
not modelled |
15.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 64 | c3opyH_ |
|
not modelled |
15.8 |
16 |
PDB header:transferase
Chain: H: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 65 | c6c02B_ |
|
not modelled |
15.6 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 3 (enpp3,2 npp3, cd203c), inactive (t205a), n594s, with alpha,beta-methylene-atp3 (ampcpp)
|
|
|
| 66 | c2dg2D_ |
|
not modelled |
15.4 |
14 |
PDB header:protein binding
Chain: D: PDB Molecule:apolipoprotein a-i binding protein;
PDBTitle: crystal structure of mouse apolipoprotein a-i binding protein
|
|
|
| 67 | c3q3qA_ |
|
not modelled |
15.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
|
|
|
| 68 | c4fdiA_ |
|
not modelled |
15.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylgalactosamine-6-sulfatase;
PDBTitle: the molecular basis of mucopolysaccharidosis iv a
|
|
|
| 69 | c3ed4A_ |
|
not modelled |
14.7 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:arylsulfatase;
PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
|
|
|
| 70 | d1fsua_ |
|
not modelled |
14.6 |
11 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 71 | c4ug4H_ |
|
not modelled |
14.4 |
10 |
PDB header:hydrolase
Chain: H: PDB Molecule:choline sulfatase;
PDBTitle: crystal structure of a choline sulfatase from sinorhizobium2 melliloti
|
|
|
| 72 | c3o3xA_ |
|
not modelled |
14.2 |
24 |
PDB header:viral protein
Chain: A: PDB Molecule:gp41-5;
PDBTitle: crystal structure of gp41-5, a single-chain 5-helix-bundle based on2 hiv gp41
|
|
|
| 73 | c3b5qB_ |
|
not modelled |
13.7 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of a putative sulfatase (np_810509.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.40 a resolution
|
|
|
| 74 | c2ponA_ |
|
not modelled |
13.0 |
28 |
PDB header:apoptosis inhibitor
Chain: A: PDB Molecule:beclin-1;
PDBTitle: solution structure of the bcl-xl/beclin-1 complex
|
|
|
| 75 | c5jp6A_ |
|
not modelled |
12.3 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative polysaccharide deacetylase;
PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
|
|
|
| 76 | d2gqfa1 |
|
not modelled |
12.2 |
57 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
|
|
| 77 | d1kxpd3 |
|
not modelled |
12.2 |
26 |
Fold:Serum albumin-like
Superfamily:Serum albumin-like
Family:Serum albumin-like |
|
|
| 78 | c3lxqB_ |
|
not modelled |
12.1 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein vp1736;
PDBTitle: the crystal structure of a protein in the alkaline phosphatase2 superfamily from vibrio parahaemolyticus to 1.95a
|
|
|
| 79 | d1m2da_ |
|
not modelled |
12.0 |
24 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
|
|
| 80 | c2w8dB_ |
|
not modelled |
11.8 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:processed glycerol phosphate lipoteichoic acid synthase 2;
PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
|
|
|
| 81 | c2iucB_ |
|
not modelled |
11.8 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:alkaline phosphatase;
PDBTitle: structure of alkaline phosphatase from the antarctic bacterium tab5
|
|
|
| 82 | c2xrgA_ |
|
not modelled |
11.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
|
|
| 83 | c5xhbA_ |
|
not modelled |
11.7 |
45 |
PDB header:immune system
Chain: A: PDB Molecule:nisin immunity protein;
PDBTitle: crystal structure of the full length of nisi in a lipid free form, the2 nisin immunity protein, from lactococcus lactis
|
|
|
| 84 | d1hdha_ |
|
not modelled |
11.3 |
15 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 85 | c4oydB_ |
|
not modelled |
11.2 |
25 |
PDB header:viral protein/inhibitor
Chain: B: PDB Molecule:computationally designed inhibitor;
PDBTitle: crystal structure of a computationally designed inhibitor of an2 epstein-barr viral bcl-2 protein
|
|
|
| 86 | d1p49a_ |
|
not modelled |
11.1 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 87 | c2m6rA_ |
|
not modelled |
10.6 |
19 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: apo_yqca
|
|
|
| 88 | c3k8wA_ |
|
not modelled |
10.4 |
20 |
PDB header:structural protein
Chain: A: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c45
|
|
|
| 89 | c2xr9A_ |
|
not modelled |
10.3 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2)
|
|
|
| 90 | c3o8nA_ |
|
not modelled |
10.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase, muscle type;
PDBTitle: structure of phosphofructokinase from rabbit skeletal muscle
|
|
|
| 91 | d1v10a3 |
|
not modelled |
10.1 |
60 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 92 | d2i09a1 |
|
not modelled |
10.1 |
18 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:DeoB catalytic domain-like |
|
|
| 93 | d1l5oa_ |
|
not modelled |
10.0 |
18 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
|
|
| 94 | c2jmkA_ |
|
not modelled |
9.8 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:hypothetical protein ta0956;
PDBTitle: solution structure of ta0956
|
|
|
| 95 | c1ihqA_ |
|
not modelled |
9.7 |
31 |
PDB header:de novo protein
Chain: A: PDB Molecule:chimeric peptide glytm1bzip: tropomyosin alpha
PDBTitle: glytm1bzip: a chimeric peptide model of the n-terminus of a2 rat short alpha tropomyosin with the n-terminus encoded by3 exon 1b
|
|
|
| 96 | c4uplC_ |
|
not modelled |
9.6 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:sulfatase family protein;
PDBTitle: dimeric sulfatase spas2 from silicibacter pomeroyi
|
|
|
| 97 | c5fqlA_ |
|
not modelled |
9.5 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:iduronate-2-sulfatase;
PDBTitle: insights into hunter syndrome from the structure of iduronate-2-2 sulfatase
|
|
|
| 98 | c3opyG_ |
|
not modelled |
9.5 |
13 |
PDB header:transferase
Chain: G: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 99 | d1j33a_ |
|
not modelled |
9.4 |
21 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
|
|




























































































































































































































































