1 c5yioA_
100.0
100
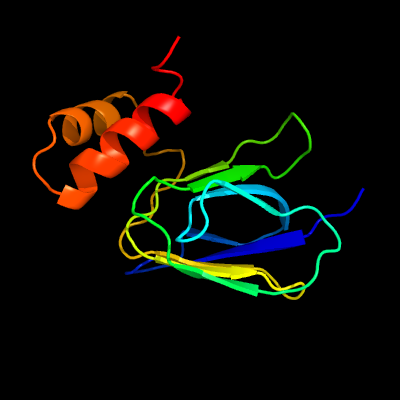
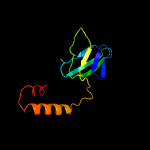
PDB header: proton transportChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: nmr solution structure of subunit epsilon of the mycobacterium2 tuberculosis f-atp synthase
2 c6q45P_
100.0
24
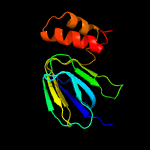
PDB header: hydrolaseChain: P: PDB Molecule: atp synthase epsilon chain;PDBTitle: f1-atpase from fusobacterium nucleatum
3 c2e5yA_
100.0
38
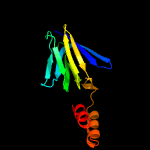
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: epsilon subunit and atp complex of f1f0-atp synthase from2 the thermophilic bacillus ps3
4 c2rq7A_
100.0
36
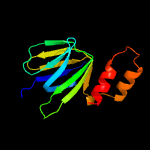
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: solution structure of the epsilon subunit chimera combining2 the n-terminal beta-sandwich domain from t. elongatus bp-13 f1 and the c-terminal alpha-helical domain from spinach4 chloroplast f1
5 c6fkhe_
100.0
32
PDB header: membrane proteinChain: E: PDB Molecule: atp synthase subunit alpha, chloroplastic;PDBTitle: chloroplast f1fo conformation 2
6 c1fs0E_
100.0
28
PDB header: hydrolaseChain: E: PDB Molecule: atp synthase epsilon subunit;PDBTitle: complex of gamma/epsilon atp synthase from e.coli
7 c6f5dH_
100.0
22
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase subunit delta, mitochondrial;PDBTitle: trypanosoma brucei f1-atpase
8 c2qe7H_
100.0
37
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase subunit epsilon;PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
9 c6refR_
100.0
21
PDB header: proton transportChain: R: PDB Molecule: mitochondrial atp synthase subunit delta;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 3b,2 monomer-masked refinement
10 c2wssQ_
100.0
27
PDB header: hydrolaseChain: Q: PDB Molecule: atp synthase subunit delta, mitochondrial;PDBTitle: the structure of the membrane extrinsic region of bovine atp synthase
11 c5lqzH_
100.0
23
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase delta subunit;PDBTitle: structure of f-atpase from pichia angusta, state1
12 c2hldH_
100.0
18
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase delta chain, mitochondrial;PDBTitle: crystal structure of yeast mitochondrial f1-atpase
13 d1aqta2
99.9
25
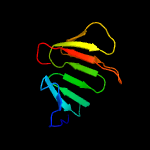
Fold: Epsilon subunit of F1F0-ATP synthase N-terminal domainSuperfamily: Epsilon subunit of F1F0-ATP synthase N-terminal domainFamily: Epsilon subunit of F1F0-ATP synthase N-terminal domain
14 c1h8eH_
99.9
22
PDB header: hydrolaseChain: H: PDB Molecule: bovine mitochondrial f1-atpase;PDBTitle: (adp.alf4)2(adp.so4) bovine f1-atpase2 (all three catalytic sites occupied)
15 c5dn6I_
99.9
27
PDB header: hydrolaseChain: I: PDB Molecule: atp synthase epsilon chain;PDBTitle: atp synthase from paracoccus denitrificans
16 c2w6jH_
97.7
27
PDB header: hydrolaseChain: H: PDB Molecule: f1-atpase delta subunit;PDBTitle: low resolution structures of bovine mitochondrial f1-atpase during2 controlled dehydration: hydration state 5.
17 c2e5tA_
91.2
27
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: c-terminal domain of epsilon subunit of f1f0-atp synthase2 from the thermophilic bacillus ps3 in the presence of atp3 condition
18 d1aqta1
78.5
39
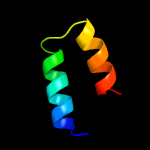
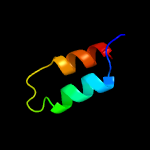
Fold: Long alpha-hairpinSuperfamily: Epsilon subunit of F1F0-ATP synthase C-terminal domainFamily: Epsilon subunit of F1F0-ATP synthase C-terminal domain
19 c3hqxA_
59.2
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0345 protein aciad0356;PDBTitle: crystal structure of protein of unknown function (duf1255,pf06865)2 from acinetobacter sp. adp1
20 c4cv7A_
23.3
16
PDB header: toxinChain: A: PDB Molecule: virulence associated protein vapb;PDBTitle: crystal structure of rhodococcus equi vapb
21 c3vtoR_
not modelled
18.6
16
PDB header: metal binding proteinChain: R: PDB Molecule: protein gp45;PDBTitle: the crystal structure of the c-terminal domain of mu phage central2 spike
22 d2ezla_
not modelled
18.2
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain
23 c4er8A_
not modelled
12.7
22
PDB header: dna binding protein/dnaChain: A: PDB Molecule: tnparep for protein;PDBTitle: structure of the rep associates tyrosine transposase bound to a rep2 hairpin
24 d1y60a_
not modelled
12.1
13
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Formaldehyde-activating enzyme, FAE
25 c3eo6B_
not modelled
8.8
5
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function (duf1255);PDBTitle: crystal structure of protein of unknown function (duf1255) (afe_2634)2 from acidithiobacillus ferrooxidans ncib8455 at 0.97 a resolution
26 c4c5eH_
not modelled
8.5
25
PDB header: transcriptionChain: H: PDB Molecule: polycomb protein pho;PDBTitle: crystal structure of the minimal pho-sfmbt complex (p21 spacegroup)
27 c4c5eG_
not modelled
8.5
25
PDB header: transcriptionChain: G: PDB Molecule: polycomb protein pho;PDBTitle: crystal structure of the minimal pho-sfmbt complex (p21 spacegroup)
28 c2vhgB_
not modelled
8.4
24
PDB header: dna-binding proteinChain: B: PDB Molecule: transposase orfa;PDBTitle: crystal structure of the ishp608 transposase in complex2 with right end 31-mer dna
29 c4gdxB_
not modelled
8.4
14
PDB header: hydrolaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase 1 light chain;PDBTitle: crystal structure of human gamma-glutamyl transpeptidase--glutamate2 complex
30 c3j6vR_
not modelled
8.1
56
PDB header: ribosomeChain: R: PDB Molecule: 28s ribosomal protein s18a, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
31 d2oyza1
not modelled
7.7
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: VPA0057-like
32 d2f5ga1
not modelled
7.3
22
Fold: Ferredoxin-likeSuperfamily: Transposase IS200-likeFamily: Transposase IS200-like
33 c4l6uB_
not modelled
6.8
27
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of af1868: cmr1 subunit of the cmr rna silencing2 complex
34 c3frnA_
not modelled
6.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flagellar protein flga;PDBTitle: crystal structure of flagellar protein flga from thermotoga maritima2 msb8
35 d1od5a1
not modelled
6.4
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein
36 c2vicA_
not modelled
6.2
24
PDB header: dna-binding proteinChain: A: PDB Molecule: transposase orfa;PDBTitle: crystal structure of the ishp608 transposase in complex2 with left end 26-mer dna and manganese
37 c5dudA_
not modelled
6.1
14
PDB header: unknown functionChain: A: PDB Molecule: ybgk;PDBTitle: crystal structure of e. coli ybgjk
38 c3j20G_
not modelled
6.0
14
PDB header: ribosomeChain: G: PDB Molecule: 30s ribosomal protein s6e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
39 c5aeoA_
not modelled
6.0
13
PDB header: immune systemChain: A: PDB Molecule: r. equi vapg protein;PDBTitle: virulence-associated protein vapg from the intracellular2 pathogen rhodococcus equi
40 d2fyxa1
not modelled
5.9
12
Fold: Ferredoxin-likeSuperfamily: Transposase IS200-likeFamily: Transposase IS200-like
41 d2vjva1
not modelled
5.9
24
Fold: Ferredoxin-likeSuperfamily: Transposase IS200-likeFamily: Transposase IS200-like
42 d1e3ha5
not modelled
5.7
22
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like
43 c5wlyA_
not modelled
5.4
20
PDB header: hydrolaseChain: A: PDB Molecule: udp-2,3-diacylglucosamine hydrolase;PDBTitle: e. coli lpxh- 8 mutations
44 c3stjC_
not modelled
5.4
11
PDB header: hydrolaseChain: C: PDB Molecule: protease degq;PDBTitle: crystal structure of the protease + pdz1 domain of degq from2 escherichia coli
45 c4bkwA_
not modelled
5.4
21
PDB header: receptorChain: A: PDB Molecule: zinc finger fyve domain-containing protein 9;PDBTitle: crystal structure of the c-terminal region of human zfyve9
46 c1tr8A_
not modelled
5.3
19
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
47 c3pm9A_
not modelled
5.3
29
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution