| 1 | c3r38A_
|
|
 |
100.0 |
48 |
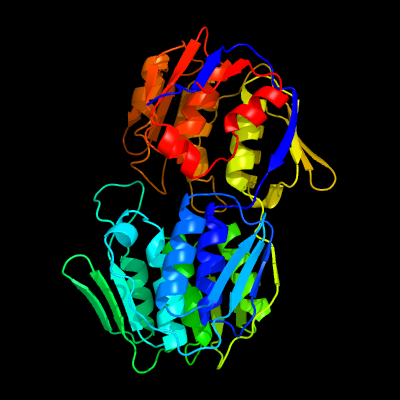
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.23 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase (mura) from listeria monocytogenes egd-e
|
|
|
|
| 2 | c5u4hB_
|
|
 |
100.0 |
46 |
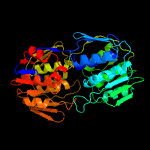
PDB header:lipid-binding protein
Chain: B: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: 1.05 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase from acinetobacter baumannii in covalently3 bound complex with (2r)-2-(phosphonooxy)propanoic acid.
|
|
|
|
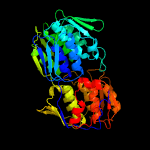
| 3 | c5wi5C_
|
|
 |
100.0 |
47 |
PDB header:transferase
Chain: C: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.0 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase from streptococcus pneumoniae in complex3 with uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2r)-4 2-(phosphonooxy)propanoic acid and magnesium.
|
|
|
|
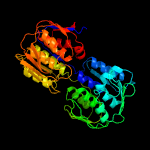
| 4 | c5ujsB_
|
|
 |
100.0 |
40 |
PDB header:hydrolase,oxidoreductase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: 2.45 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase from campylobacter jejuni.
|
|
|
|
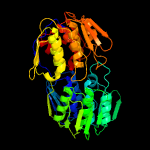
| 5 | d1uaea_
|
|
 |
100.0 |
45 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
|
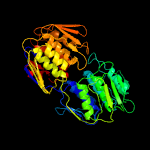
| 6 | c5bq2C_
|
|
 |
100.0 |
44 |
PDB header:transferase
Chain: C: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: crystal structure of udp-n-acetylglucosamine 1-carboxyvinyltransferase2 (udp-n-acetylglucosamine enolpyruvyl transferase, ept) from3 pseudomonas aeruginosa
|
|
|
|
| 7 | d1ejda_
|
|
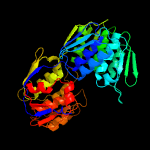 |
100.0 |
44 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
|
| 8 | c2yvwA_
|
|
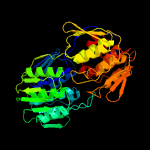 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: crystal structure of udp-n-acetylglucosamine 1-carboxyvinyltransferase2 from aquifex aeolicus vf5
|
|
|
|
| 9 | c3zh3A_
|
|
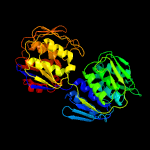 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: crystal structure of s. pneumoniae d39 native mura1
|
|
|
|
| 10 | c4fqdA_
|
|
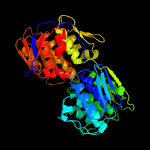 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:niko protein;
PDBTitle: crystal structure of the enolpyruvyl transferase niko from2 streptomyces tendae
|
|
|
|
| 11 | c3rmtB_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase 1;
PDBTitle: crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate2 synthase from bacillus halodurans c-125
|
|
|
|
| 12 | d1g6sa_
|
|
 |
100.0 |
20 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
|
| 13 | c3roiA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: 2.20 angstrom resolution structure of 3-phosphoshikimate 1-2 carboxyvinyltransferase (aroa) from coxiella burnetii
|
|
|
|
| 14 | c5xwbB_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: crystal structure of 5-enolpyruvulshikimate-3-phosphate synthase from2 a psychrophilic bacterium, colwellia psychrerythraea
|
|
|
|
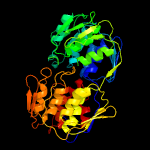
| 15 | c2o0zA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: mycobacterium tuberculosis epsp synthase in complex with product (eps)
|
|
|
|
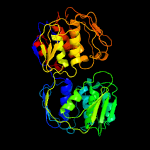
| 16 | c5bufA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: 2.37 angstrom structure of epsp synthase from acinetobacter baumannii
|
|
|
|
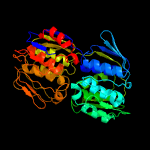
| 17 | c2pqdA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: a100g cp4 epsps liganded with (r)-difluoromethyl tetrahedral reaction2 intermediate analog
|
|
|
|
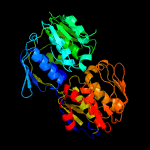
| 18 | d1rf6a_
|
|
 |
100.0 |
23 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
|
| 19 | d1p88a_
|
|
 |
100.0 |
19 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
|
| 20 | d1qmha2
|
|
 |
97.3 |
18 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:RNA 3'-terminal phosphate cyclase, RPTC |
|
|
|
| 21 | c1qmiC_ |
|
not modelled |
95.4 |
16 |
PDB header:ligase
Chain: C: PDB Molecule:rna 3'-terminal phosphate cyclase;
PDBTitle: crystal structure of rna 3'-terminal phosphate cyclase, an2 ubiquitous enzyme with unusual topology
|
|
|
| 22 | c4o8jB_ |
|
not modelled |
95.2 |
18 |
PDB header:ligase/rna
Chain: B: PDB Molecule:rna 3'-terminal phosphate cyclase;
PDBTitle: crystal structure of rtca, the rna 3'-terminal phosphate cyclase from2 pyrococcus horikoshii, in complex with racaaa3'phosphate and adenine.
|
|
|
| 23 | c3pqvD_ |
|
not modelled |
85.4 |
12 |
PDB header:unknown function
Chain: D: PDB Molecule:rcl1 protein;
PDBTitle: cyclase homolog
|
|
|
| 24 | c3g0tA_ |
|
not modelled |
56.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aspartate aminotransferase (np_905498.1)2 from porphyromonas gingivalis w83 at 1.75 a resolution
|
|
|
| 25 | c4gtnA_ |
|
not modelled |
47.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: structure of anthranilate phosphoribosyl transferase from2 acinetobacter baylyi
|
|
|
| 26 | c1o17A_ |
|
not modelled |
45.3 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
|
|
|
| 27 | c4muoB_ |
|
not modelled |
43.5 |
10 |
PDB header:dna binding protein
Chain: B: PDB Molecule:uncharacterized protein ybib;
PDBTitle: the trpd2 enzyme from e.coli: ybib
|
|
|
| 28 | c2hzfA_ |
|
not modelled |
40.7 |
16 |
PDB header:electron transport, oxidoreductase
Chain: A: PDB Molecule:glutaredoxin-1;
PDBTitle: crystal structures of a poxviral glutaredoxin in the oxidized and2 reduced states show redox-correlated structural changes
|
|
|
| 29 | d1w2za3 |
|
not modelled |
37.7 |
19 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
|
|
| 30 | c3gmgB_ |
|
not modelled |
25.0 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein rv1825/mt1873;
PDBTitle: crystal structure of an uncharacterized conserved protein from2 mycobacterium tuberculosis
|
|
|
| 31 | c2dsjA_ |
|
not modelled |
21.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside (thymidine) phosphorylase;
PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
|
|
|
| 32 | c1v8gB_ |
|
not modelled |
20.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase2 (trpd) from thermus thermophilus hb8
|
|
|
| 33 | c5kinD_ |
|
not modelled |
19.6 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 34 | c4kjeA_ |
|
not modelled |
17.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin;
PDBTitle: atomic resolution structure of pfgrx1
|
|
|
| 35 | c3pnxF_ |
|
not modelled |
16.5 |
0 |
PDB header:transferase
Chain: F: PDB Molecule:putative sulfurtransferase dsre;
PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
|
|
|
| 36 | c4j8cA_ |
|
not modelled |
14.8 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:hsc70-interacting protein;
PDBTitle: crystal structure of the dimerization domain of hsc70-interacting2 protein
|
|
|
| 37 | c4j8cB_ |
|
not modelled |
14.8 |
23 |
PDB header:chaperone
Chain: B: PDB Molecule:hsc70-interacting protein;
PDBTitle: crystal structure of the dimerization domain of hsc70-interacting2 protein
|
|
|
| 38 | c2bpqB_ |
|
not modelled |
14.1 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase (trpd) from2 mycobacterium tuberculosis (apo structure)
|
|
|
| 39 | d1v8za1 |
|
not modelled |
14.1 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
|
|
| 40 | c4ga5H_ |
|
not modelled |
13.7 |
13 |
PDB header:transferase
Chain: H: PDB Molecule:putative thymidine phosphorylase;
PDBTitle: crystal structure of amp phosphorylase c-terminal deletion mutant in2 the apo-form
|
|
|
| 41 | d2o8ra3 |
|
not modelled |
13.3 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 42 | c5fqlA_ |
|
not modelled |
13.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:iduronate-2-sulfatase;
PDBTitle: insights into hunter syndrome from the structure of iduronate-2-2 sulfatase
|
|
|
| 43 | d1o17a2 |
|
not modelled |
12.9 |
10 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 44 | c2cb1A_ |
|
not modelled |
12.8 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
|
|
| 45 | d1jjcb2 |
|
not modelled |
12.4 |
24 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Domains B1 and B5 of PheRS-beta, PheT |
|
|
| 46 | c3juxA_ |
|
not modelled |
12.0 |
8 |
PDB header:protein transport
Chain: A: PDB Molecule:protein translocase subunit seca;
PDBTitle: structure of the translocation atpase seca from thermotoga2 maritima
|
|
|
| 47 | c3h5qA_ |
|
not modelled |
12.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
|
|
| 48 | c3nr6A_ |
|
not modelled |
11.9 |
13 |
PDB header:hydrolase/inhibitor
Chain: A: PDB Molecule:protease p14;
PDBTitle: crystal structure of xenotropic murine leukemia virus-related virus2 (xmrv) protease
|
|
|
| 49 | c4i2uA_ |
|
not modelled |
10.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin;
PDBTitle: crystal structure of the reduced glutaredoxin from chlorella2 sorokiniana t-89 in complex with glutathione
|
|
|
| 50 | d1jhba_ |
|
not modelled |
10.7 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
|
|
| 51 | c3dinB_ |
|
not modelled |
10.7 |
8 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:protein translocase subunit seca;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
|
|
|
| 52 | c2gjhA_ |
|
not modelled |
9.7 |
29 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein;
PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
|
|
|
| 53 | c5tchH_ |
|
not modelled |
9.6 |
20 |
PDB header:lyase
Chain: H: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 54 | c1khdD_ |
|
not modelled |
9.5 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.9 resolution3 (current name, pectobacterium carotovorum)
|
|
|
| 55 | d1nh2a1 |
|
not modelled |
9.1 |
18 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 56 | d2dy1a4 |
|
not modelled |
9.1 |
24 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
|
|
| 57 | d1lxja_ |
|
not modelled |
9.0 |
11 |
Fold:Ferredoxin-like
Superfamily:MTH1187/YkoF-like
Family:MTH1187-like |
|
|
| 58 | d1cdwa1 |
|
not modelled |
8.7 |
11 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 59 | c5nofB_ |
|
not modelled |
8.2 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase from thermococcus kodakaraensis
|
|
|
| 60 | d1in0a2 |
|
not modelled |
7.5 |
21 |
Fold:Ferredoxin-like
Superfamily:YajQ-like
Family:YajQ-like |
|
|
| 61 | d1qnaa1 |
|
not modelled |
7.3 |
11 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 62 | c2o8rA_ |
|
not modelled |
7.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
|
|
|
| 63 | c1nl3B_ |
|
not modelled |
6.8 |
17 |
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase seca 1 subunit;
PDBTitle: crystal structure of the seca protein translocation atpase2 from mycobacterium tuberculosis in apo form
|
|
|
| 64 | d1x9za_ |
|
not modelled |
6.8 |
27 |
Fold:DNA mismatch repair protein MutL
Superfamily:DNA mismatch repair protein MutL
Family:DNA mismatch repair protein MutL |
|
|
| 65 | c4pwyA_ |
|
not modelled |
6.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:calmodulin-lysine n-methyltransferase;
PDBTitle: crystal structure of a calmodulin-lysine n-methyltransferase fragment
|
|
|
| 66 | d1yqha1 |
|
not modelled |
6.7 |
23 |
Fold:Ferredoxin-like
Superfamily:MTH1187/YkoF-like
Family:MTH1187-like |
|
|
| 67 | c3ndnC_ |
|
not modelled |
6.6 |
22 |
PDB header:lyase
Chain: C: PDB Molecule:o-succinylhomoserine sulfhydrylase;
PDBTitle: crystal structure of o-succinylhomoserine sulfhydrylase from2 mycobacterium tuberculosis covalently bound to pyridoxal-5-phosphate
|
|
|
| 68 | c2cxiA_ |
|
not modelled |
6.5 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:phenylalanyl-trna synthetase beta chain;
PDBTitle: crystal structure of an n-terminal fragment of the phenylalanyl-trna2 synthetase beta-subunit from pyrococcus horikoshii
|
|
|
| 69 | c6ic4H_ |
|
not modelled |
6.4 |
23 |
PDB header:protein transport
Chain: H: PDB Molecule:abc transporter permease;
PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
|
|
|
| 70 | c2epiA_ |
|
not modelled |
6.1 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0045 protein mj1052;
PDBTitle: crystal structure pf hypothetical protein mj1052 from2 methanocaldococcus jannascii (form 2)
|
|
|
| 71 | c5zvlB_ |
|
not modelled |
5.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutaredoxin;
PDBTitle: crystal structure of wheat glutarredoxin
|
|
|
| 72 | c3mjsA_ |
|
not modelled |
5.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:amphb;
PDBTitle: structure of a-type ketoreductases from modular polyketide synthase
|
|
|
| 73 | c3nbxX_ |
|
not modelled |
5.9 |
12 |
PDB header:hydrolase
Chain: X: PDB Molecule:atpase rava;
PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
|
|
|
| 74 | d1nh8a2 |
|
not modelled |
5.7 |
14 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain |
|
|
| 75 | c4gduB_ |
|
not modelled |
5.6 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of sulfate-bound human l-asparaginase protein
|
|
|
| 76 | c4uaqA_ |
|
not modelled |
5.6 |
11 |
PDB header:protein transport
Chain: A: PDB Molecule:protein translocase subunit seca 2;
PDBTitle: crystal structure of the accessory translocation atpase, seca2, from2 mycobacterium tuberculosis
|
|
|
| 77 | d1ttea1 |
|
not modelled |
5.5 |
24 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:UBA domain |
|
|
| 78 | c4n0bA_ |
|
not modelled |
5.3 |
15 |
PDB header:transcription activator
Chain: A: PDB Molecule:hth-type transcriptional regulatory protein gabr;
PDBTitle: crystal structure of bacillus subtilis gabr, an autorepressor and2 transcriptional activator of gabt
|
|
|
| 79 | c1otpA_ |
|
not modelled |
5.2 |
16 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
|
|
| 80 | c3h8qB_ |
|
not modelled |
5.2 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 3;
PDBTitle: crystal structure of glutaredoxin domain of human thioredoxin2 reductase 3
|
|
|
| 81 | c3s8iA_ |
|
not modelled |
5.2 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein ddi1 homolog 1;
PDBTitle: the retroviral-like protease (rvp) domain of human ddi1
|
|
|
| 82 | c2k6xA_ |
|
not modelled |
5.2 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
|
|
|
| 83 | c3ke2A_ |
|
not modelled |
5.1 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein yp_928783.1;
PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
|
|
|