1 c2kduB_
62.3
27
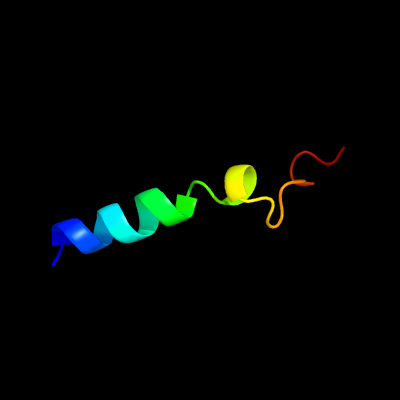
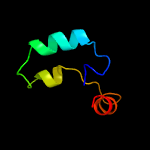
PDB header: metal binding protein/exocytosisChain: B: PDB Molecule: protein unc-13 homolog a;PDBTitle: structural basis of the munc13-1/ca2+-calmodulin2 interaction: a novel 1-26 calmodulin binding motif with a3 bipartite binding mode
2 c6hqaF_
23.0
31
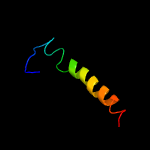
PDB header: transcriptionChain: F: PDB Molecule: subunit (17 kda) of tfiid and saga complexes, involved inPDBTitle: molecular structure of promoter-bound yeast tfiid
3 c2ebkA_
23.0
24
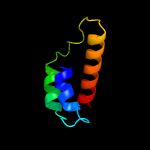
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: rwd domain-containing protein 3;PDBTitle: solution structure of the rwd domain of human rwd domain2 containing protein 3
4 d1wgqa_
20.9
18
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
5 d2dawa1
20.1
18
Fold: UBC-likeSuperfamily: UBC-likeFamily: RWD domain
6 d1qqga1
18.0
27
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Phosphotyrosine-binding domain (PTB)
7 d2elba2
16.5
27
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
8 c5j1hB_
15.6
24
PDB header: structural proteinChain: B: PDB Molecule: plectin,plectin;PDBTitle: structure of the spectrin repeats 5 and 6 of the plakin domain of2 plectin
9 c5todD_
13.3
44
PDB header: lipid transportChain: D: PDB Molecule: transmembrane protein 24;PDBTitle: transmembrane protein 24 smp domain
10 d2daxa1
13.1
8
Fold: UBC-likeSuperfamily: UBC-likeFamily: RWD domain
11 c2jo7A_
12.3
38
PDB header: surface active proteinChain: A: PDB Molecule: glycosylphosphatidylinositol-anchored merozoitePDBTitle: solution structure of the adhesion protein bd37 from2 babesia divergens
12 c5hwiA_
11.5
38
PDB header: transferaseChain: A: PDB Molecule: glutathione-specific gamma-glutamylcyclotransferase;PDBTitle: crystal structure of selenomethionine labelled gama glutamyl2 cyclotransferease specific to glutathione from yeast
13 d1x1ga1
10.9
27
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
14 d1u5fa1
10.7
23
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
15 d1ddfa_
10.5
11
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD
16 c2wsc1_
10.0
22
PDB header: photosynthesisChain: 1: PDB Molecule: at3g54890;PDBTitle: improved model of plant photosystem i
17 c3nsuA_
9.6
13
PDB header: signaling proteinChain: A: PDB Molecule: phosphatidylinositol 4,5-bisphosphate-binding protein slm1;PDBTitle: a systematic screen for protein-lipid interactions in saccharomyces2 cerevisiae
18 c3cqcA_
9.4
19
PDB header: protein transportChain: A: PDB Molecule: nuclear pore complex protein nup107;PDBTitle: nucleoporin nup107/nup133 interaction complex
19 c2ws94_
9.4
60
PDB header: virusChain: 4: PDB Molecule: p1;PDB Fragment: capsid protein vp4, residues 1-80
PDBTitle: equine rhinitis a virus at low ph
20 c3fybA_
9.4
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein of unknown function (duf1244);PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
21 c4cwlv_
not modelled
9.2
60
PDB header: virusChain: V: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
22 c4cwlx_
not modelled
9.2
60
PDB header: virusChain: X: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
23 c4cwlI_
not modelled
9.2
60
PDB header: virusChain: I: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
24 c4cwlo_
not modelled
9.2
60
PDB header: virusChain: O: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
25 c4cwlq_
not modelled
9.2
60
PDB header: virusChain: Q: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
26 c4cwlG_
not modelled
9.2
60
PDB header: virusChain: G: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
27 c4cwlK_
not modelled
9.2
60
PDB header: virusChain: K: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
28 c4cwlB_
not modelled
9.2
60
PDB header: virusChain: B: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
29 c4cwl2_
not modelled
9.2
60
PDB header: virusChain: 2: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
30 c4cwlD_
not modelled
9.2
60
PDB header: virusChain: D: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
31 c4cwlf_
not modelled
9.2
60
PDB header: virusChain: F: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
32 c4cwlb_
not modelled
9.2
60
PDB header: virusChain: B: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
33 c4cwl7_
not modelled
9.2
60
PDB header: virusChain: 7: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
34 c4cwlk_
not modelled
9.2
60
PDB header: virusChain: K: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
35 c4cwl9_
not modelled
9.2
60
PDB header: virusChain: 9: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
36 c4cwlg_
not modelled
9.2
60
PDB header: virusChain: G: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
37 c4cwl0_
not modelled
9.2
60
PDB header: virusChain: 0: PDB Molecule: PDB Fragment: residues 1-80
PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
38 c4cwl8_
not modelled
9.2
60
PDB header: virusChain: 8: PDB Molecule: PDB Fragment: residues 1-80
PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
39 c4cwlF_
not modelled
9.2
60
PDB header: virusChain: F: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
40 c4cwlA_
not modelled
9.2
60
PDB header: virusChain: A: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
41 c4cwlu_
not modelled
9.2
60
PDB header: virusChain: U: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
42 c4cwlM_
not modelled
9.2
60
PDB header: virusChain: M: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
43 c4cwlV_
not modelled
9.2
60
PDB header: virusChain: V: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
44 c4cwlm_
not modelled
9.2
60
PDB header: virusChain: M: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
45 c4cwlS_
not modelled
9.2
60
PDB header: virusChain: S: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
46 c4cwlN_
not modelled
9.2
60
PDB header: virusChain: N: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
47 c4cwlU_
not modelled
9.2
60
PDB header: virusChain: U: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
48 c4cwlL_
not modelled
9.2
60
PDB header: virusChain: L: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
49 c4cwli_
not modelled
9.2
60
PDB header: virusChain: I: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
50 c4cwlX_
not modelled
9.2
60
PDB header: virusChain: X: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
51 c4cwlt_
not modelled
9.2
60
PDB header: virusChain: T: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
52 c4cwl5_
not modelled
9.2
60
PDB header: virusChain: 5: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
53 c4cwlp_
not modelled
9.2
60
PDB header: virusChain: P: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
54 c4cwld_
not modelled
9.2
60
PDB header: virusChain: D: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
55 c4cwle_
not modelled
9.2
60
PDB header: virusChain: E: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
56 c4cwlQ_
not modelled
9.2
60
PDB header: virusChain: Q: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
57 c4cwlr_
not modelled
9.2
60
PDB header: virusChain: R: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
58 c4cwlP_
not modelled
9.2
60
PDB header: virusChain: P: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
59 c4cwlw_
not modelled
9.2
60
PDB header: virusChain: W: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
60 c4cwlH_
not modelled
9.2
60
PDB header: virusChain: H: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
61 c4cwlZ_
not modelled
9.2
60
PDB header: virusChain: Z: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
62 c4cwlC_
not modelled
9.2
60
PDB header: virusChain: C: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
63 c4cwlT_
not modelled
9.2
60
PDB header: virusChain: T: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
64 c4cwlO_
not modelled
9.2
60
PDB header: virusChain: O: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
65 c4cwl4_
not modelled
9.2
60
PDB header: virusChain: 4: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
66 c4cwlc_
not modelled
9.2
60
PDB header: virusChain: C: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
67 c4cwlJ_
not modelled
9.2
60
PDB header: virusChain: J: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
68 c4cwlE_
not modelled
9.2
60
PDB header: virusChain: E: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
69 c4cwla_
not modelled
9.2
60
PDB header: virusChain: A: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
70 c4cwlR_
not modelled
9.2
60
PDB header: virusChain: R: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
71 c4cwls_
not modelled
9.2
60
PDB header: virusChain: S: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
72 c4cwll_
not modelled
9.2
60
PDB header: virusChain: L: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
73 c4cwl3_
not modelled
9.2
60
PDB header: virusChain: 3: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
74 c4cwlh_
not modelled
9.2
60
PDB header: virusChain: H: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
75 c4cwl6_
not modelled
9.2
60
PDB header: virusChain: 6: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
76 c4cwl1_
not modelled
9.2
60
PDB header: virusChain: 1: PDB Molecule: PDB Fragment: residues 1-80
PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
77 c4cwln_
not modelled
9.2
60
PDB header: virusChain: N: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
78 c4cwlW_
not modelled
9.2
60
PDB header: virusChain: W: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
79 c4cwlY_
not modelled
9.2
60
PDB header: virusChain: Y: PDB Molecule: PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
80 c2wff4_
not modelled
9.2
60
PDB header: virusChain: 4: PDB Molecule: p1;PDB Fragment: capsid protein vp4, residues 1-80
PDBTitle: equine rhinitis a virus
81 c4cwlj_
not modelled
9.2
60
PDB header: virusChain: J: PDB Molecule: p1;PDBTitle: the limits of structural plasticity in a picornavirus2 capsid revealed by a massively expanded equine rhinitis a3 virus particle
82 d2daya1
not modelled
9.1
23
Fold: UBC-likeSuperfamily: UBC-likeFamily: RWD domain
83 d1u5da1
not modelled
9.0
20
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
84 c2xbo4_
not modelled
8.9
60
PDB header: virusChain: 4: PDB Molecule: p1;PDB Fragment: capsid protein vp4, residues 1-80
PDBTitle: equine rhinitis a virus in complex with its sialic acid2 receptor
85 d1ukxa_
not modelled
8.9
17
Fold: UBC-likeSuperfamily: UBC-likeFamily: RWD domain
86 c2dhiA_
not modelled
8.2
31
PDB header: signaling proteinChain: A: PDB Molecule: pleckstrin homology domain-containing family bPDBTitle: solution structure of the ph domain of evectin-2 from mouse
87 c1wmvA_
not modelled
8.1
15
PDB header: oxidoreductase, apoptosisChain: A: PDB Molecule: ww domain containing oxidoreductase;PDBTitle: solution structure of the second ww domain of wwox
88 c3pp2A_
not modelled
8.0
20
PDB header: hydrolase activatorChain: A: PDB Molecule: rho gtpase-activating protein 27;PDBTitle: crystal structure of the pleckstrin homology domain of arhgap27
89 d2i5ua1
not modelled
7.9
19
Fold: DnaD domain-likeSuperfamily: DnaD domain-likeFamily: DnaD domain
90 d1ie6a_
not modelled
7.7
56
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
91 d1u29a1
not modelled
7.5
20
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
92 c1u5fA_
not modelled
7.4
20
PDB header: signaling proteinChain: A: PDB Molecule: src-associated adaptor protein;PDBTitle: crystal structure of the ph domain of skap-hom with 8 vector-derived2 n-terminal residues
93 c3enoC_
not modelled
7.4
16
PDB header: hydrolase/unknown functionChain: C: PDB Molecule: uncharacterized protein pf2011;PDBTitle: crystal structure of pyrococcus furiosus pcc1 in complex2 with thermoplasma acidophilum kae1
94 d1l0wa2
not modelled
7.3
18
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain
95 d1tafa_
not modelled
7.2
46
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs
96 d1unqa_
not modelled
6.9
21
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
97 d1ghea_
not modelled
6.5
27
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
98 d1fgya_
not modelled
6.4
7
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
99 c2mcfA_
not modelled
6.4
78
PDB header: unknown functionChain: A: PDB Molecule: tgam_1934;PDBTitle: nmr structure of tgam_1934