1 c3s3uB_
100.0
52
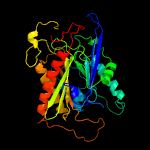
PDB header: transferaseChain: B: PDB Molecule: cysteine transferase;PDBTitle: crystal structure of uncleaved thnt t282c
2 c3n5iC_
100.0
26
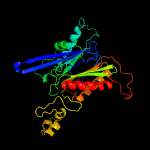
PDB header: hydrolaseChain: C: PDB Molecule: beta-peptidyl aminopeptidase;PDBTitle: crystal structure of the precursor (s250a mutant) of the n-terminal2 beta-aminopeptidase bapa
3 d1b65a_
100.0
24
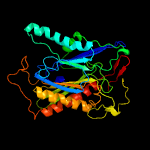
Fold: DmpA/ArgJ-likeSuperfamily: DmpA/ArgJ-likeFamily: DmpA-like
4 c2drhD_
100.0
25
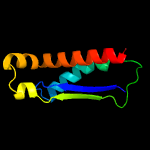
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: 361aa long hypothetical d-aminopeptidase;PDBTitle: crystal structure of the ph0078 protein from pyrococcus horikoshii ot3
5 c3axgN_
100.0
31
PDB header: hydrolaseChain: N: PDB Molecule: endotype 6-aminohexanoat-oligomer hydrolase;PDBTitle: structure of 6-aminohexanoate-oligomer hydrolase
6 c5tzbA_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: d-aminopeptidase;PDBTitle: burkholderia sp. beta-aminopeptidase
7 c3it4B_
95.3
29
PDB header: transferaseChain: B: PDB Molecule: arginine biosynthesis bifunctional protein argjPDBTitle: the crystal structure of ornithine acetyltransferase from2 mycobacterium tuberculosis (rv1653) at 1.7 a
8 d1vz6a_
95.2
19
Fold: DmpA/ArgJ-likeSuperfamily: DmpA/ArgJ-likeFamily: ArgJ-like
9 c1vraB_
94.3
15
PDB header: transferaseChain: B: PDB Molecule: arginine biosynthesis bifunctional protein argj;PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
10 c2vzkD_
92.6
20
PDB header: transferaseChain: D: PDB Molecule: glutamate n-acetyltransferase 2 beta chain;PDBTitle: structure of the acyl-enzyme complex of an n-terminal nucleophile2 (ntn) hydrolase, oat2
11 c4qmkB_
87.8
38
PDB header: toxinChain: B: PDB Molecule: type iii secretion system effector protein exou;PDBTitle: crystal structure of type iii effector protein exou (exou)
12 c3tu3B_
83.2
42
PDB header: toxin/toxin chaperoneChain: B: PDB Molecule: exou;PDBTitle: 1.92 angstrom resolution crystal structure of the full-length spcu in2 complex with full-length exou from the type iii secretion system of3 pseudomonas aeruginosa
13 c4akxB_
70.7
38
PDB header: transport proteinChain: B: PDB Molecule: exou;PDBTitle: structure of the heterodimeric complex exou-spcu from the type iii2 secretion system (t3ss) of pseudomonas aeruginosa
14 c5fyaA_
54.5
35
PDB header: hydrolaseChain: A: PDB Molecule: patatin-like protein, plpd;PDBTitle: cubic crystal of the native plpd
15 c4akfA_
50.3
32
PDB header: transferaseChain: A: PDB Molecule: vipd;PDBTitle: crystal structure of vipd from legionella pneumophila
16 c3geiB_
29.8
40
PDB header: hydrolaseChain: B: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
17 c2m4mA_
19.4
10
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: solution structure of the rrm domain of the hypothetical protein2 cagl0m09691g from candida glabrata
18 c2jpiA_
17.6
19
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
19 c2vg2C_
15.2
20
PDB header: transferaseChain: C: PDB Molecule: undecaprenyl pyrophosphate synthetase;PDBTitle: rv2361 with ipp
20 c1osxA_
14.5
54
PDB header: immune systemChain: A: PDB Molecule: tumor necrosis factor receptor superfamilyPDBTitle: solution structure of the extracellular domain of blys2 receptor 3 (br3)
21 d1osxa_
not modelled
14.5
54
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like
22 c1p0tA_
not modelled
14.4
54
PDB header: protein bindingChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
23 c1p0tU_
not modelled
14.4
54
PDB header: protein bindingChain: U: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
24 c1p0tx_
not modelled
14.4
54
PDB header: protein bindingChain: X: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
25 c1p0tN_
not modelled
14.4
54
PDB header: protein bindingChain: N: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
26 c1p0tC_
not modelled
14.4
54
PDB header: protein bindingChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
27 c1p0tk_
not modelled
14.4
54
PDB header: protein bindingChain: K: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
28 c1p0t3_
not modelled
14.4
54
PDB header: protein bindingChain: 3: PDB Molecule: PDB Fragment: residues 1-63;
PDBTitle: crystal structure of the baff-baff-r complex (part ii)
29 c1p0tl_
not modelled
14.4
54
PDB header: protein bindingChain: L: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
30 c1p0tV_
not modelled
14.4
54
PDB header: protein bindingChain: V: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
31 c1p0tW_
not modelled
14.4
54
PDB header: protein bindingChain: W: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
32 c1p0tp_
not modelled
14.4
54
PDB header: protein bindingChain: P: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
33 c1p0tX_
not modelled
14.4
54
PDB header: protein bindingChain: X: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
34 c1p0t9_
not modelled
14.4
54
PDB header: protein bindingChain: 9: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
35 c1p0tJ_
not modelled
14.4
54
PDB header: protein bindingChain: J: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
36 c1p0tB_
not modelled
14.4
54
PDB header: protein bindingChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
37 d1qy9a1
not modelled
14.3
22
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
38 c1p0tK_
not modelled
14.1
54
PDB header: protein bindingChain: K: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
39 c1p0te_
not modelled
14.1
54
PDB header: protein bindingChain: E: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
40 c1p0t7_
not modelled
14.1
54
PDB header: protein bindingChain: 7: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
41 c1p0t0_
not modelled
14.1
54
PDB header: protein bindingChain: 0: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
42 c1p0tZ_
not modelled
14.1
54
PDB header: protein bindingChain: Z: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
43 c1p0tt_
not modelled
14.1
54
PDB header: protein bindingChain: T: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
44 c1p0tO_
not modelled
14.1
54
PDB header: protein bindingChain: O: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
45 c1p0tq_
not modelled
14.1
54
PDB header: protein bindingChain: Q: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
46 c1p0tn_
not modelled
14.1
54
PDB header: protein bindingChain: N: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
47 c1p0t6_
not modelled
14.1
54
PDB header: protein bindingChain: 6: PDB Molecule: PDB Fragment: residues 1-63;
PDBTitle: crystal structure of the baff-baff-r complex (part ii)
48 c1p0tF_
not modelled
14.1
54
PDB header: protein bindingChain: F: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
49 c1p0t1_
not modelled
14.1
54
PDB header: protein bindingChain: 1: PDB Molecule: PDB Fragment: residues 1-63;
PDBTitle: crystal structure of the baff-baff-r complex (part ii)
50 c1p0t8_
not modelled
14.1
54
PDB header: protein bindingChain: 8: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
51 c1p0tf_
not modelled
14.1
54
PDB header: protein bindingChain: F: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
52 c1p0tR_
not modelled
14.1
54
PDB header: protein bindingChain: R: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
53 c1p0ta_
not modelled
14.1
54
PDB header: protein bindingChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
54 c1p0tw_
not modelled
14.1
54
PDB header: protein bindingChain: W: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
55 c1p0ts_
not modelled
14.1
54
PDB header: protein bindingChain: S: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
56 c1p0ti_
not modelled
14.1
54
PDB header: protein bindingChain: I: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
57 c1p0td_
not modelled
14.1
54
PDB header: protein bindingChain: D: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
58 c1p0th_
not modelled
14.1
54
PDB header: protein bindingChain: H: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
59 c1p0tT_
not modelled
14.1
54
PDB header: protein bindingChain: T: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
60 c1p0tE_
not modelled
14.1
54
PDB header: protein bindingChain: E: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
61 c1p0tM_
not modelled
14.1
54
PDB header: protein bindingChain: M: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
62 c1p0tm_
not modelled
14.1
54
PDB header: protein bindingChain: M: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
63 c1p0t4_
not modelled
14.1
54
PDB header: protein bindingChain: 4: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
64 c1p0tH_
not modelled
14.1
54
PDB header: protein bindingChain: H: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
65 c1p0tI_
not modelled
14.1
54
PDB header: protein bindingChain: I: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
66 c1p0tr_
not modelled
14.1
54
PDB header: protein bindingChain: R: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
67 c1p0to_
not modelled
14.1
54
PDB header: protein bindingChain: O: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
68 c1p0tS_
not modelled
14.1
54
PDB header: protein bindingChain: S: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
69 c1p0tv_
not modelled
14.1
54
PDB header: protein bindingChain: V: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
70 c1p0tY_
not modelled
14.1
54
PDB header: protein bindingChain: Y: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
71 c1p0t5_
not modelled
14.1
54
PDB header: protein bindingChain: 5: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
72 c1p0tj_
not modelled
14.1
54
PDB header: protein bindingChain: J: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
73 c1p0tP_
not modelled
14.1
54
PDB header: protein bindingChain: P: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
74 c1p0tc_
not modelled
14.1
54
PDB header: protein bindingChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
75 c1p0tD_
not modelled
14.1
54
PDB header: protein bindingChain: D: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
76 c1p0tG_
not modelled
14.1
54
PDB header: protein bindingChain: G: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
77 c1p0t2_
not modelled
14.1
54
PDB header: protein bindingChain: 2: PDB Molecule: PDBTitle: crystal structure of the baff-baff-r complex (part ii)
78 c1p0tb_
not modelled
14.1
54
PDB header: protein bindingChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
79 c1p0tL_
not modelled
14.1
54
PDB header: protein bindingChain: L: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
80 c1p0tg_
not modelled
14.1
54
PDB header: protein bindingChain: G: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
81 c1p0tQ_
not modelled
14.1
54
PDB header: protein bindingChain: Q: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
82 c1p0tu_
not modelled
14.1
54
PDB header: protein bindingChain: U: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of the baff-baff-r complex (part ii)
83 d1oqen_
not modelled
13.9
54
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like
84 c1oqeN_
not modelled
13.9
54
PDB header: immune systemChain: N: PDB Molecule: tumor necrosis factor receptor superfamily member 13c;PDBTitle: crystal structure of stall-1 with baff-r
85 c1pjtB_
not modelled
13.5
12
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
86 d1xuba1
not modelled
13.1
25
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
87 c4m55E_
not modelled
12.3
20
PDB header: lyaseChain: E: PDB Molecule: udp-glucuronic acid decarboxylase 1;PDBTitle: crystal structure of human udp-xylose synthase r236h substitution
88 c4yn3B_
not modelled
10.5
21
PDB header: hydrolaseChain: B: PDB Molecule: cucumisin;PDBTitle: crystal structure of cucumisin complex with pro-peptide
89 c5uazB_
not modelled
9.7
50
PDB header: protein transportChain: B: PDB Molecule: nucleoporin nup53;PDBTitle: crystal structure of the yeast nucleoporin
90 d1ws6a1
not modelled
9.1
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like
91 c1kyqC_
not modelled
9.0
16
PDB header: oxidoreductase, lyaseChain: C: PDB Molecule: siroheme biosynthesis protein met8;PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
92 c4u3fV_
not modelled
8.9
26
PDB header: oxidoreductase/oxidoreductase inhibitorChain: V: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: cytochrome bc1 complex from chicken with designed inhibitor bound
93 c5gukA_
not modelled
8.7
27
PDB header: biosynthetic proteinChain: A: PDB Molecule: cyclolavandulyl diphosphate synthase;PDBTitle: crystal structure of apo form of cyclolavandulyl diphosphate synthase2 (clds) from streptomyces sp. cl190
94 c3h1kI_
not modelled
8.6
20
PDB header: oxidoreductaseChain: I: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: chicken cytochrome bc1 complex with zn++ and an iodinated derivative2 of kresoxim-methyl bound
95 c3sxpD_
not modelled
8.2
15
PDB header: isomeraseChain: D: PDB Molecule: adp-l-glycero-d-mannoheptose-6-epimerase;PDBTitle: crystal structure of helicobacter pylori adp-l-glycero-d-manno-2 heptose-6-epimerase (rfad, hp0859)
96 c3h1lV_
not modelled
7.8
26
PDB header: oxidoreductaseChain: V: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: chicken cytochrome bc1 complex with ascochlorin bound at qo and qi2 sites
97 c3cwbV_
not modelled
7.5
26
PDB header: oxidoreductaseChain: V: PDB Molecule: mitochondrial ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
98 d2o0ma1
not modelled
7.5
21
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like
99 c2o0mA_
not modelled
7.5
21
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, sorc family;PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis