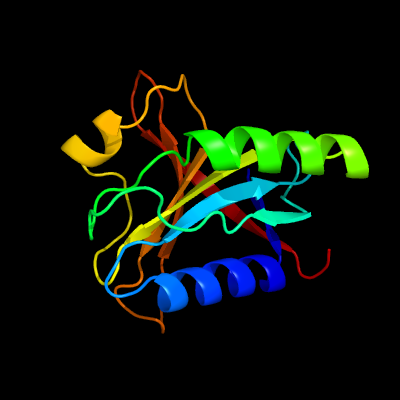
| 1 | c2kksA_
|
|
 |
100.0 |
34 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein dsy2949 from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr27
|
|
|
|
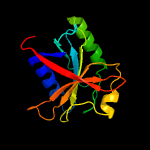
| 2 | c2kcqA_
|
|
 |
100.0 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mov34/mpn/pad-1 family;
PDBTitle: solution structure of protein sru_2040 from salinibacter2 ruber (strain dsm 13855) . northeast structural genomics3 consortium target srr106
|
|
|
|
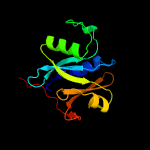
| 3 | c5ld9B_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:jamm1;
PDBTitle: structure of deubiquitinating enzyme homolog, pyrococcus furiosus2 jamm1.
|
|
|
|
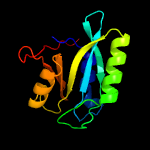
| 4 | c5cw6A_
|
|
 |
100.0 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:drbrcc36;
PDBTitle: structure of metal dependent enzyme drbrcc36
|
|
|
|
| 5 | d1oi0a_
|
|
 |
99.9 |
24 |
Fold:Cytidine deaminase-like
Superfamily:JAB1/MPN domain
Family:JAB1/MPN domain |
|
|
|
| 6 | c6fnnB_
|
|
 |
99.9 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:26s proteasome regulatory subunit n11-like protein;
PDBTitle: caldiarchaeum subterraneum ubiquitin:rpn11-homolog complex
|
|
|
|
| 7 | c6r8fA_
|
|
 |
99.9 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:lys-63-specific deubiquitinase brcc36;
PDBTitle: cryo-em structure of the human brisc-shmt2 complex
|
|
|
|
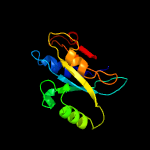
| 8 | c6fjuA_
|
|
 |
99.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:26s proteasome regulatory subunit n11-like protein;
PDBTitle: rpn11 homolog from caldiarchaeum subterraneum
|
|
|
|
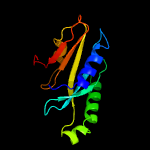
| 9 | c5cw4C_
|
|
 |
99.9 |
16 |
PDB header:metal binding protein
Chain: C: PDB Molecule:brca1/brca2-containing complex subunit 3;
PDBTitle: structure of cfbrcc36-cfkiaa0157 complex (selenium edge)
|
|
|
|
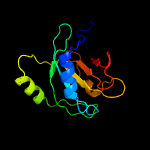
| 10 | c4b4tV_
|
|
 |
99.9 |
23 |
PDB header:hydrolase
Chain: V: PDB Molecule:26s proteasome regulatory subunit rpn11;
PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
|
|
|
|
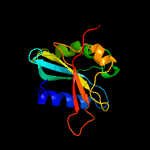
| 11 | c4d18E_
|
|
 |
99.9 |
19 |
PDB header:signaling protein
Chain: E: PDB Molecule:cop9 signalosome complex subunit 5;
PDBTitle: crystal structure of the cop9 signalosome
|
|
|
|
| 12 | c5gjqV_
|
|
 |
99.9 |
21 |
PDB header:hydrolase
Chain: V: PDB Molecule:26s proteasome non-atpase regulatory subunit 14;
PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
|
|
|
|
| 13 | c2znrA_
|
|
 |
99.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:amsh-like protease;
PDBTitle: crystal structure of the dub domain of human amsh-lp
|
|
|
|
| 14 | c4k1rA_
|
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:amsh-like protease sst2;
PDBTitle: crystal structure of schizosaccharomyces pombe sst2 catalytic domain2 and ubiquitin
|
|
|
|
| 15 | c4f7oB_
|
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:cop9 signalosome complex subunit 5;
PDBTitle: crystal structure of csn5
|
|
|
|
| 16 | c3j7kH_
|
|
 |
99.7 |
18 |
PDB header:translation
Chain: H: PDB Molecule:eukaryotic translation initiation factor 3 subunit h;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
|
|
|
|
| 17 | c3j7jH_
|
|
 |
99.7 |
18 |
PDB header:translation
Chain: H: PDB Molecule:eukaryotic translation initiation factor 3 subunit h;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
|
|
|
|
| 18 | c3j8cH_
|
|
 |
99.7 |
18 |
PDB header:translation
Chain: H: PDB Molecule:eukaryotic translation initiation factor 3 subunit h;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
|
|
|
|
| 19 | c3j8bH_
|
|
 |
99.7 |
18 |
PDB header:translation
Chain: H: PDB Molecule:eukaryotic translation initiation factor 3 subunit h;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
|
|
|
|
| 20 | c5a5tH_
|
|
 |
99.6 |
17 |
PDB header:hydrolase
Chain: H: PDB Molecule:eukaryotic translation initiation factor 3 subunit h;
PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
|
|
|
|
| 21 | c2p87A_ |
|
not modelled |
99.4 |
17 |
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-splicing factor prp8;
PDBTitle: crystal structure of the c-terminal domain of c. elegans2 pre-mrna splicing factor prp8
|
|
|
| 22 | c4kitC_ |
|
not modelled |
99.4 |
15 |
PDB header:rna binding protein
Chain: C: PDB Molecule:pre-mrna-processing-splicing factor 8;
PDBTitle: crystal structure of human brr2 in complex with the prp8 jab1/mpn2 domain
|
|
|
| 23 | c2og4A_ |
|
not modelled |
99.4 |
11 |
PDB header:protein binding
Chain: A: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: structure of an expanded jab1-mpn-like domain of splicing factor prp8p2 from yeast
|
|
|
| 24 | c2o95A_ |
|
not modelled |
99.3 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:26s proteasome non-atpase regulatory subunit 7;
PDBTitle: crystal structure of the metal-free dimeric human mov34 mpn domain2 (residues 1-186)
|
|
|
| 25 | c5m59F_ |
|
not modelled |
99.3 |
15 |
PDB header:splicing
Chain: F: PDB Molecule:putative pre-mrna splicing factor;
PDBTitle: crystal structure of chaetomium thermophilum brr2 helicase core in2 complex with prp8 jab1 domain
|
|
|
| 26 | c3sbgA_ |
|
not modelled |
99.3 |
12 |
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: crystal structure of a prp8 c-terminal fragment
|
|
|
| 27 | c4cr4U_ |
|
not modelled |
99.3 |
16 |
PDB header:hydrolase
Chain: U: PDB Molecule:26s proteasome regulatory subunit rpn8;
PDBTitle: deep classification of a large cryo-em dataset defines the2 conformational landscape of the 26s proteasome
|
|
|
| 28 | c4b4tU_ |
|
not modelled |
99.2 |
16 |
PDB header:hydrolase
Chain: U: PDB Molecule:26s proteasome regulatory subunit rpn8;
PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
|
|
|
| 29 | c4d18F_ |
|
not modelled |
99.2 |
10 |
PDB header:signaling protein
Chain: F: PDB Molecule:cop9 signalosome complex subunit 6;
PDBTitle: crystal structure of the cop9 signalosome
|
|
|
| 30 | c5a5tF_ |
|
not modelled |
99.1 |
14 |
PDB header:hydrolase
Chain: F: PDB Molecule:eukaryotic translation initiation factor 3 subunit f;
PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
|
|
|
| 31 | c5gjqU_ |
|
not modelled |
99.1 |
15 |
PDB header:hydrolase
Chain: U: PDB Molecule:26s proteasome non-atpase regulatory subunit 7;
PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
|
|
|
| 32 | c3j8bF_ |
|
not modelled |
98.9 |
14 |
PDB header:translation
Chain: F: PDB Molecule:eukaryotic translation initiation factor 3 subunit f;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
|
|
|
| 33 | c3j7kF_ |
|
not modelled |
98.9 |
14 |
PDB header:translation
Chain: F: PDB Molecule:eukaryotic translation initiation factor 3 subunit f;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
|
|
|
| 34 | c3j8cF_ |
|
not modelled |
98.9 |
14 |
PDB header:translation
Chain: F: PDB Molecule:eukaryotic translation initiation factor 3 subunit f;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
|
|
|
| 35 | c3j7jF_ |
|
not modelled |
98.9 |
14 |
PDB header:translation
Chain: F: PDB Molecule:eukaryotic translation initiation factor 3 subunit f;
PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
|
|
|
| 36 | c4e0qB_ |
|
not modelled |
98.9 |
11 |
PDB header:unknown function
Chain: B: PDB Molecule:cop9 signalosome complex subunit 6;
PDBTitle: crystal structure of mpn domain from cop9 signalosome
|
|
|
| 37 | c5m32n_ |
|
not modelled |
98.9 |
16 |
PDB header:hydrolase
Chain: N: PDB Molecule:proteasome subunit beta type-6;
PDBTitle: human 26s proteasome in complex with oprozomib
|
|
|
| 38 | c6cddB_ |
|
not modelled |
97.6 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:npl4 zinc finger;
PDBTitle: npl4 zinc finger and mpn domains (chaetomium thermophilum)
|
|
|
| 39 | c4i43B_ |
|
not modelled |
97.4 |
13 |
PDB header:splicing
Chain: B: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: crystal structure of prp8:aar2 complex
|
|
|
| 40 | c5z58A_ |
|
not modelled |
97.4 |
18 |
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-processing-splicing factor 8;
PDBTitle: cryo-em structure of a human activated spliceosome (early bact) at 4.92 angstrom.
|
|
|
| 41 | c6oa9G_ |
|
not modelled |
97.4 |
13 |
PDB header:motor protein
Chain: G: PDB Molecule:nuclear protein localization protein 4;
PDBTitle: cdc48-npl4 complex processing poly-ubiquitinated substrate in the2 presence of atp
|
|
|
| 42 | c3jcrA_ |
|
not modelled |
97.4 |
17 |
PDB header:splicing
Chain: A: PDB Molecule:hprp8;
PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
|
|
|
| 43 | c6oaaG_ |
|
not modelled |
97.3 |
14 |
PDB header:motor protein
Chain: G: PDB Molecule:nuclear protein localization protein 4;
PDBTitle: cdc48-npl4 complex processing poly-ubiquitinated substrate in the2 presence of adp-befx, state 1
|
|
|
| 44 | c5lj5A_ |
|
not modelled |
97.0 |
15 |
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: overall structure of the yeast spliceosome immediately after2 branching.
|
|
|
| 45 | c6gvwF_ |
|
not modelled |
96.9 |
16 |
PDB header:signaling protein
Chain: F: PDB Molecule:brca1-a complex subunit abraxas 1;
PDBTitle: crystal structure of the brca1-a complex
|
|
|
| 46 | c5ganA_ |
|
not modelled |
96.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
|
|
|
| 47 | c5lqwA_ |
|
not modelled |
96.4 |
15 |
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-splicing factor 8;
PDBTitle: yeast activated spliceosome
|
|
|
| 48 | c6r8fD_ |
|
not modelled |
96.4 |
15 |
PDB header:signaling protein
Chain: D: PDB Molecule:brisc complex subunit abraxas 2;
PDBTitle: cryo-em structure of the human brisc-shmt2 complex
|
|
|
| 49 | c5cw5D_ |
|
not modelled |
95.9 |
11 |
PDB header:metal binding protein
Chain: D: PDB Molecule:protein fam175b;
PDBTitle: structure of cfbrcc36-cfkiaa0157 complex (qsq mutant)
|
|
|
| 50 | c2qlcC_ |
|
not modelled |
95.4 |
13 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna repair protein radc homolog;
PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
|
|
|
| 51 | d1knwa2 |
|
not modelled |
53.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
|
|
| 52 | c5bwaA_ |
|
not modelled |
39.6 |
7 |
PDB header:lyase/lyase inhibitor
Chain: A: PDB Molecule:ornithine decarboxylase;
PDBTitle: crystal structure of odc-plp-az1 ternary complex
|
|
|
| 53 | c1knwA_ |
|
not modelled |
25.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase
|
|
|
| 54 | c1w8gA_ |
|
not modelled |
22.8 |
11 |
PDB header:plp-binding protein
Chain: A: PDB Molecule:hypothetical upf0001 protein yggs;
PDBTitle: crystal structure of e. coli k-12 yggs
|
|
|
| 55 | c5nm8A_ |
|
not modelled |
21.5 |
7 |
PDB header:plp-binding protein
Chain: A: PDB Molecule:pipy;
PDBTitle: structure of pipy, the cog0325 family member of synechococcus2 elongatus pcc7942, with plp bound
|
|
|
| 56 | d1gska1 |
|
not modelled |
20.0 |
38 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 57 | c2qghA_ |
|
not modelled |
19.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from helicobacter2 pylori complexed with l-lysine
|
|
|
| 58 | d1x87a_ |
|
not modelled |
19.0 |
10 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
|
|
| 59 | c6hbeA_ |
|
not modelled |
17.4 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:copper-containing nitrite reductase;
PDBTitle: cu-containing nitrite reductase (nirk) from thermus scotoductus sa-01
|
|
|
| 60 | c3aw5A_ |
|
not modelled |
17.3 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:multicopper oxidase;
PDBTitle: structure of a multicopper oxidase from the hyperthermophilic archaeon2 pyrobaculum aerophilum
|
|
|
| 61 | c1njjC_ |
|
not modelled |
17.2 |
6 |
PDB header:lyase
Chain: C: PDB Molecule:ornithine decarboxylase;
PDBTitle: crystal structure determination of t. brucei ornithine decarboxylase2 bound to d-ornithine and to g418
|
|
|
| 62 | c6evgA_ |
|
not modelled |
16.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:multi-copper oxidase cueo;
PDBTitle: structural and functional characterisation of a bacterial laccase-like2 multi-copper oxidase cueo from lignin-degrading bacterium3 ochrobactrum sp. with oxidase activity towards lignin model compounds4 and lignosulfonate
|
|
|
| 63 | d7odca2 |
|
not modelled |
16.8 |
3 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
|
|
| 64 | c2fknC_ |
|
not modelled |
16.4 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:urocanate hydratase;
PDBTitle: crystal structure of urocanase from bacillus subtilis
|
|
|
| 65 | c3zx1A_ |
|
not modelled |
16.2 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, putative;
PDBTitle: multicopper oxidase from campylobacter jejuni: a metallo-oxidase
|
|
|
| 66 | d2isya2 |
|
not modelled |
16.0 |
8 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
|
|
| 67 | c3cdzA_ |
|
not modelled |
15.5 |
60 |
PDB header:blood clotting
Chain: A: PDB Molecule:coagulation factor viii heavy chain;
PDBTitle: crystal structure of human factor viii
|
|
|
| 68 | d1uwka_ |
|
not modelled |
15.5 |
17 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
|
|
| 69 | d2j5wa4 |
|
not modelled |
14.0 |
23 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 70 | d1g01a_ |
|
not modelled |
14.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 71 | c5zl1A_ |
|
not modelled |
13.9 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative copper-type nitrite reductase;
PDBTitle: hexameric structure of copper-containing nitrite reductase of an2 anammox organism ksu-1
|
|
|
| 72 | c4xg1C_ |
|
not modelled |
13.8 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: psychromonas ingrahamii diaminopimelate decarboxylase with llp
|
|
|
| 73 | c5gjmB_ |
|
not modelled |
13.7 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:lysine/ornithine decarboxylase;
PDBTitle: crystal strcuture of lysine decarboxylase from selenomonas ruminantium2 in c2 space group
|
|
|
| 74 | d1sddb1 |
|
not modelled |
13.5 |
15 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 75 | c4kbxA_ |
|
not modelled |
13.2 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein yhfx;
PDBTitle: crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx2 from escherichia coli
|
|
|
| 76 | c4eyzB_ |
|
not modelled |
13.2 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:cellulosome-related protein module from ruminococcus
PDBTitle: crystal structure of an uncommon cellulosome-related protein module2 from ruminococcus flavefaciens that resembles papain-like cysteine3 peptidases
|
|
|
| 77 | c3abgA_ |
|
not modelled |
13.0 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bilirubin oxidase;
PDBTitle: x-ray crystal analysis of bilirubin oxidase from myrothecium2 verrucaria at 2.3 angstrom resolution using a twin crystal
|
|
|
| 78 | c4eclA_ |
|
not modelled |
12.5 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:serine racemase;
PDBTitle: crystal structure of the cytoplasmic domain of vancomycin resistance2 serine racemase vantg
|
|
|
| 79 | c2fqeA_ |
|
not modelled |
12.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:blue copper oxidase cueo;
PDBTitle: crystal structures of e. coli laccase cueo under different copper2 binding situations
|
|
|
| 80 | d1mzya1 |
|
not modelled |
12.4 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 81 | d1kv7a1 |
|
not modelled |
12.4 |
22 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 82 | c2g23G_ |
|
not modelled |
12.3 |
60 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:phenoxazinone synthase;
PDBTitle: the crystal structure of hexameric phenoxazinone synthase
|
|
|
| 83 | c3n2bD_ |
|
not modelled |
12.0 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
|
|
|
| 84 | c5l20A_ |
|
not modelled |
11.4 |
16 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:clostripain-related protein;
PDBTitle: crystal structure of a clostripain (bt_0727) from bacteroides2 thetaiotaomicron atcc 29148 in complex with peptide inhibitor btn-3 vltk-aomk
|
|
|
| 85 | c1zpuE_ |
|
not modelled |
11.2 |
80 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:iron transport multicopper oxidase fet3;
PDBTitle: crystal structure of fet3p, a multicopper oxidase that functions in2 iron import
|
|
|
| 86 | c3ppsD_ |
|
not modelled |
11.2 |
80 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:laccase;
PDBTitle: crystal structure of an ascomycete fungal laccase from thielavia2 arenaria
|
|
|
| 87 | c1wa1X_ |
|
not modelled |
11.1 |
22 |
PDB header:reductase
Chain: X: PDB Molecule:dissimilatory copper-containing nitrite reductase;
PDBTitle: crystal structure of h313q mutant of alcaligenes xylosoxidans nitrite2 reductase
|
|
|
| 88 | c2xllC_ |
|
not modelled |
11.0 |
38 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:bilirubin oxidase;
PDBTitle: the crystal structure of bilirubin oxidase from myrothecium2 verrucaria
|
|
|
| 89 | d2v94a1 |
|
not modelled |
10.9 |
57 |
Fold:Ribosomal proteins S24e, L23 and L15e
Superfamily:Ribosomal proteins S24e, L23 and L15e
Family:Ribosomal protein S24e |
|
|
| 90 | c2q9oA_ |
|
not modelled |
10.7 |
80 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:laccase-1;
PDBTitle: near-atomic resolution structure of a melanocarpus albomyces laccase
|
|
|
| 91 | c3izbU_ |
|
not modelled |
10.5 |
21 |
PDB header:ribosome
Chain: U: PDB Molecule:40s ribosomal protein s24;
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 92 | c5lwxA_ |
|
not modelled |
9.4 |
80 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:multicopper oxidase;
PDBTitle: crystal structure of the h253d mutant of mcog from aspergillus niger
|
|
|
| 93 | d1sdda1 |
|
not modelled |
8.9 |
38 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 94 | d2q9oa1 |
|
not modelled |
8.9 |
80 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 95 | d1hfua1 |
|
not modelled |
8.8 |
80 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|
| 96 | c1mzzC_ |
|
not modelled |
8.7 |
8 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:copper-containing nitrite reductase;
PDBTitle: crystal structure of mutant (m182t)of nitrite reductase
|
|
|
| 97 | c5dynA_ |
|
not modelled |
8.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative peptidase;
PDBTitle: b. fragilis cysteine protease
|
|
|
| 98 | c1gycA_ |
|
not modelled |
8.6 |
80 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:laccase 2;
PDBTitle: crystal structure determination at room temperature of a laccase from2 trametes versicolor in its oxidised form containing a full complement3 of copper ions
|
|
|
| 99 | d1aoza1 |
|
not modelled |
8.5 |
60 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
|
|