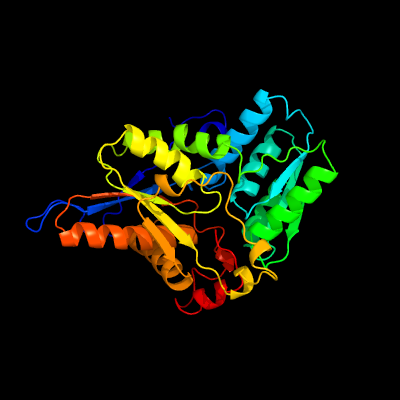
1 c3dwgA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase b;PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
2 c5ohxB_
100.0
36
PDB header: lyaseChain: B: PDB Molecule: cystathionine beta-synthase;PDBTitle: structure of active cystathionine b-synthase from apis mellifera
3 c3pc3A_
100.0
35
PDB header: lyaseChain: A: PDB Molecule: cg1753, isoform a;PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
4 c4l3vB_
100.0
34
PDB header: lyaseChain: B: PDB Molecule: cystathionine beta-synthase;PDBTitle: crystal structure of delta516-525 human cystathionine beta-synthase
5 d1jbqa_
100.0
35
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
6 c1jbqD_
100.0
35
PDB header: lyaseChain: D: PDB Molecule: cystathionine beta-synthase;PDBTitle: structure of human cystathionine beta-synthase: a unique pyridoxal 5'-2 phosphate dependent hemeprotein
7 c6c2qA_
100.0
35
PDB header: lyaseChain: A: PDB Molecule: cystathionine beta-synthase;PDBTitle: crystal structures of cystathionine beta-synthase from saccharomyces2 cerevisiae: the structure of the plp-l-serine intermediate
8 c2pqmA_
100.0
34
PDB header: lyaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
9 c4aecB_
100.0
36
PDB header: lyaseChain: B: PDB Molecule: cysteine synthase, mitochondrial;PDBTitle: crystal structure of the arabidopsis thaliana o-acetyl-serine-(thiol)-2 lyase c
10 c3x43F_
100.0
40
PDB header: transferaseChain: F: PDB Molecule: o-ureido-l-serine synthase;PDBTitle: crystal structure of o-ureido-l-serine synthase
11 d1z7wa1
100.0
36
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
12 c5d87A_
100.0
36
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable siderophore biosynthesis protein sbna;PDBTitle: staphyloferrin b precursor biosynthetic enzyme sbna y152f/s185g2 variant
13 c4lmaA_
100.0
34
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure analysis of o-acetylserine sulfhydrylase cysk1 from2 microcystis aeruginosa 7806
14 c5b1iC_
100.0
35
PDB header: lyaseChain: C: PDB Molecule: cystathionine beta-synthase;PDBTitle: crystal structure of k42a mutant of cystathionine beta-synthase from2 lactobacillus plantarum in a complex with l-methionine
15 c3vbeA_
100.0
37
PDB header: transferaseChain: A: PDB Molecule: beta-cyanoalnine synthase;PDBTitle: crystal structure of beta-cyanoalanine synthase in soybean
16 c4i1xA_
100.0
31
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of cysteine synthase from helicobacter pylori 26695
17 d2bhsa1
100.0
42
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
18 c4airB_
100.0
35
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: leishmania major cysteine synthase
19 c5i7wA_
100.0
33
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase a;PDBTitle: crystal structure of a cysteine synthase from brucella suis
20 c5xa2B_
100.0
39
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of o-acetylserine sulfhydrylase from planctomyces2 limnophila
21 c4ql4A_
not modelled
100.0
38
PDB header: lyaseChain: A: PDB Molecule: o-acetylserine lyase;PDBTitle: crystal structure of o-acetylserine sulfhydrylase from bacillus2 anthracis
22 c2q3bA_
not modelled
100.0
39
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase a;PDBTitle: 1.8 a resolution crystal structure of o-acetylserine sulfhydrylase2 (oass) holoenzyme from mycobacterium tuberculosis
23 d1ve1a1
not modelled
100.0
46
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
24 d1y7la1
not modelled
100.0
35
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
25 c5xenB_
not modelled
100.0
37
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of a hydrogen sulfide-producing enzyme (fn1220) from2 fusobacterium nucleatum in complex with l-serine-plp schiff base
26 d1wkva1
not modelled
100.0
29
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
27 c2eguA_
not modelled
100.0
43
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of o-acetylserine sulfhydrase from geobacillus2 kaustophilus hta426
28 d1fcja_
not modelled
100.0
38
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
29 d1o58a_
not modelled
100.0
42
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
30 c3l6cA_
not modelled
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: serine racemase;PDBTitle: x-ray crystal structure of rat serine racemase in complex with2 malonate a potent inhibitor
31 c6hulB_
not modelled
100.0
20
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase beta chain 1;PDBTitle: sulfolobus solfataricus tryptophan synthase ab complex
32 c1tdjA_
not modelled
100.0
19
PDB header: allosteryChain: A: PDB Molecule: biosynthetic threonine deaminase;PDBTitle: threonine deaminase (biosynthetic) from e. coli
33 d1pwha_
not modelled
100.0
26
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
34 c4qysA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain 2;PDBTitle: trpb2 enzymes
35 c2d1fA_
not modelled
100.0
18
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: structure of mycobacterium tuberculosis threonine synthase
36 c4negA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: the crystal structure of tryptophan synthase subunit beta from2 bacillus anthracis str. 'ames ancestor'
37 d1v71a1
not modelled
100.0
17
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
38 c2gn0A_
not modelled
100.0
16
PDB header: lyaseChain: A: PDB Molecule: threonine dehydratase catabolic;PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
39 c1x1qA_
not modelled
100.0
21
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
40 c3iauA_
not modelled
100.0
17
PDB header: lyaseChain: A: PDB Molecule: threonine deaminase;PDBTitle: the structure of the processed form of threonine deaminase isoform 22 from solanum lycopersicum
41 c5ybwA_
not modelled
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyridoxal 5'-phosphate-dependent aspartate2 racemase
42 d1tdja1
not modelled
100.0
20
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
43 c1p5jA_
not modelled
100.0
22
PDB header: lyaseChain: A: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure analysis of human serine dehydratase
44 d1p5ja_
not modelled
100.0
22
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
45 c5b54D_
not modelled
100.0
35
PDB header: transferaseChain: D: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of hydrogen sulfide-producing enzyme (fn1055) from2 fusobacterium nucleatum: lysine-dimethylated form
46 d1v8za1
not modelled
100.0
22
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
47 d1qopb_
not modelled
100.0
20
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
48 d1v7ca_
not modelled
100.0
21
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
49 c5cvcB_
not modelled
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: serine racemase;PDBTitle: structure of maize serine racemase
50 c3r0zA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of apo d-serine deaminase from salmonella2 typhimurium
51 d1ve5a1
not modelled
100.0
23
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
52 c5tchH_
not modelled
100.0
21
PDB header: lyaseChain: H: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
53 c6cgqA_
not modelled
100.0
24
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: threonine synthase from bacillus subtilis atcc 6633 with plp and plp-2 ala
54 c2o2jA_
not modelled
100.0
23
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain dimer2 (apoform)
55 c5c3uA_
not modelled
100.0
20
PDB header: lyaseChain: A: PDB Molecule: l-serine ammonia-lyase;PDBTitle: crystal structure of a fungal l-serine ammonia-lyase from rhizomucor2 miehei
56 c2zsjB_
not modelled
100.0
18
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
57 c5kinD_
not modelled
100.0
21
PDB header: lyaseChain: D: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
58 c4d9gA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: putative diaminopropionate ammonia-lyase;PDBTitle: crystal structure of selenomethionine incorporated holo2 diaminopropionate ammonia lyase from escherichia coli
59 c2rkbE_
not modelled
100.0
24
PDB header: lyaseChain: E: PDB Molecule: serine dehydratase-like;PDBTitle: serine dehydratase like-1 from human cancer cells
60 c5ygrA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: diaminopropionate ammonia lyase;PDBTitle: crystal structure of plp bound diaminopropionate ammonia lyase from2 salmonella typhimurium
61 d1e5xa_
not modelled
100.0
15
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
62 d1f2da_
not modelled
100.0
17
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
63 d1j0aa_
not modelled
100.0
17
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
64 c4d8tC_
not modelled
100.0
17
PDB header: lyaseChain: C: PDB Molecule: d-cysteine desulfhydrase;PDBTitle: crystal structure of d-cysteine desulfhydrase from salmonella2 typhimurium at 2.2 a resolution
65 d1tyza_
not modelled
100.0
19
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
66 c3v7nA_
not modelled
100.0
18
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase (thrc) from from burkholderia2 thailandensis
67 d1vb3a1
not modelled
100.0
19
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
68 c4f4fB_
not modelled
100.0
16
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: x-ray crystal structure of plp bound threonine synthase from brucella2 melitensis
69 d1kl7a_
not modelled
99.9
14
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
70 d1bg6a2
not modelled
93.8
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
71 d1c1da1
not modelled
92.8
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
72 d1kola2
not modelled
92.7
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
73 c3iupB_
not modelled
91.5
20
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadph:quinone oxidoreductase;PDBTitle: crystal structure of putative nadph:quinone oxidoreductase2 (yp_296108.1) from ralstonia eutropha jmp134 at 1.70 a resolution
74 d1vp8a_
not modelled
91.4
19
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like
75 d1o8ca2
not modelled
91.4
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
76 d1o89a2
not modelled
90.8
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
77 c3widC_
not modelled
87.4
11
PDB header: oxidoreductaseChain: C: PDB Molecule: glucose 1-dehydrogenase;PDBTitle: structure of a glucose dehydrogenase t277f mutant in complex with nadp
78 c3zu3A_
not modelled
87.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ypo4104/y4119/yp_4011;PDBTitle: structure of the enoyl-acp reductase fabv from yersinia pestis with2 the cofactor nadh (mr, cleaved histag)
79 c3s8mA_
not modelled
84.1
19
PDB header: oxidoreductaseChain: A: PDB Molecule: enoyl-acp reductase;PDBTitle: the crystal structure of fabv
80 d1llua2
not modelled
82.7
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
81 c2douA_
not modelled
82.1
11
PDB header: transferaseChain: A: PDB Molecule: probable n-succinyldiaminopimelate aminotransferase;PDBTitle: probable n-succinyldiaminopimelate aminotransferase (ttha0342) from2 thermus thermophilus hb8
82 d1ml4a2
not modelled
81.2
22
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
83 d1tt7a2
not modelled
80.7
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
84 c3p2yA_
not modelled
80.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: alanine dehydrogenase/pyridine nucleotide transhydrogenase;PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
85 c1m67A_
not modelled
78.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol-3-phosphate dehydrogenase;PDBTitle: crystal structure of leishmania mexicana gpdh complexed with inhibitor2 2-bromo-6-hydroxy-purine
86 c5ijzH_
not modelled
78.1
23
PDB header: oxidoreductaseChain: H: PDB Molecule: nadp-specific glutamate dehydrogenase;PDBTitle: crystal strcuture of glutamate dehydrogenase(gdh) from corynebacterium2 glutamicum
87 d1ykfa2
not modelled
77.1
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
88 d1cdoa2
not modelled
76.6
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
89 d1l7da1
not modelled
75.4
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
90 c4gi2B_
not modelled
75.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: crotonyl-coa carboxylase/reductase;PDBTitle: crotonyl-coa carboxylase/reductase
91 d1p0fa2
not modelled
74.9
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
92 d1rjwa2
not modelled
74.5
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
93 d1xa0a2
not modelled
73.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
94 c3krtC_
not modelled
73.8
22
PDB header: oxidoreductaseChain: C: PDB Molecule: crotonyl coa reductase;PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
95 c3r3jC_
not modelled
72.4
15
PDB header: oxidoreductaseChain: C: PDB Molecule: glutamate dehydrogenase;PDBTitle: kinetic and structural characterization of plasmodium falciparum2 glutamate dehydrogenase 2
96 c3orqA_
not modelled
72.3
23
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
97 c4k2bA_
not modelled
69.0
17
PDB header: transferaseChain: A: PDB Molecule: ntd biosynthesis operon protein ntda;PDBTitle: crystal structure of ntda from bacillus subtilis in complex with the2 internal aldimine
98 d1jq5a_
not modelled
67.0
18
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase
99 c4euhA_
not modelled
66.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ca_c0462;PDBTitle: crystal structure of clostridium acetobutulicum trans-2-enoyl-coa2 reductase apo form
100 d1hyua1
not modelled
65.4
30
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
101 d1fl2a1
not modelled
65.4
30
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
102 d1ekxa2
not modelled
64.5
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
103 c4ak9A_
not modelled
64.3
12
PDB header: protein transportChain: A: PDB Molecule: cpftsy;PDBTitle: structure of chloroplast ftsy from physcomitrella patens
104 d2q7wa1
not modelled
64.1
7
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
105 d1trba1
not modelled
64.0
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
106 d1y6ja1
not modelled
63.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
107 c5ajtA_
not modelled
63.8
11
PDB header: hydrolaseChain: A: PDB Molecule: phosphoribohydrolase lonely guy;PDBTitle: crystal structure of ligand-free phosphoribohydrolase lonely guy from2 claviceps purpurea
108 c2vq3B_
not modelled
63.7
28
PDB header: oxidoreductaseChain: B: PDB Molecule: metalloreductase steap3;PDBTitle: crystal structure of the membrane proximal oxidoreductase2 domain of human steap3, the dominant ferric reductase of3 the erythroid transferrin cycle
109 d1o69a_
not modelled
63.6
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
110 c4wd2A_
not modelled
63.1
12
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid transaminase tyrb;PDBTitle: crystal structure of an aromatic amino acid aminotransferase from2 burkholderia cenocepacia j2315
111 c3f0hA_
not modelled
60.8
10
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
112 c4effA_
not modelled
60.6
11
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid aminotransferase;PDBTitle: crystal structure of aromatic-amino-acid aminotransferase from2 burkholderia pseudomallei
113 c3getA_
not modelled
60.1
12
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (np_281508.1) from campylobacter jejuni at 2.01 a resolution
114 c2vhyB_
not modelled
59.5
14
PDB header: oxidoreductaseChain: B: PDB Molecule: alanine dehydrogenase;PDBTitle: crystal structure of apo l-alanine dehydrogenase from mycobacterium2 tuberculosis
115 c4ggoA_
not modelled
59.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: trans-2-enoyl-coa reductase;PDBTitle: crystal structure of trans-2-enoyl-coa reductase from treponema2 denticola
116 d1vdca1
not modelled
58.9
15
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
117 c3grkE_
not modelled
58.7
17
PDB header: oxidoreductaseChain: E: PDB Molecule: enoyl-(acyl-carrier-protein) reductase (nadh);PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
118 c4g56C_
not modelled
58.4
16
PDB header: transferaseChain: C: PDB Molecule: hsl7 protein;PDBTitle: crystal structure of full length prmt5/mep50 complexes from xenopus2 laevis
119 c6hnuA_
not modelled
58.3
6
PDB header: transferaseChain: A: PDB Molecule: aromatic amino acid aminotransferase i;PDBTitle: crystal structure of the aminotransferase aro8 from c. albicans with2 ligands
120 c3e2yB_
not modelled
58.0
9
PDB header: transferase, lyaseChain: B: PDB Molecule: kynurenine-oxoglutarate transaminase 3;PDBTitle: crystal structure of mouse kynurenine aminotransferase iii in complex2 with glutamine