| 1 |
|
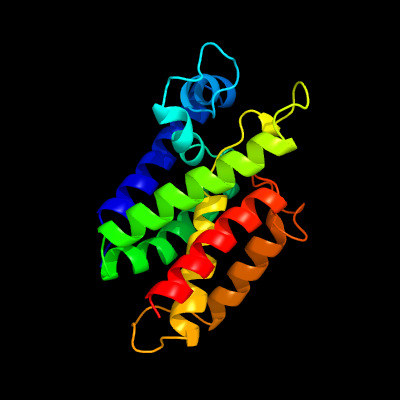
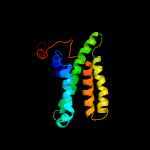
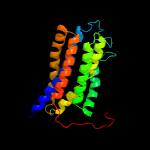
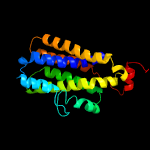
PDB 3b45 chain A domain 1
Region: 33 - 219
Aligned: 178
Modelled: 187
Confidence: 100.0%
Identity: 17%
Fold: Rhomboid-like
Superfamily: Rhomboid-like
Family: Rhomboid-like
Phyre2
| 2 |
|
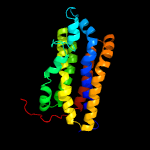
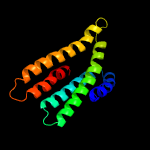
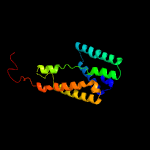
PDB 2nr9 chain A domain 1
Region: 32 - 227
Aligned: 187
Modelled: 196
Confidence: 99.9%
Identity: 14%
Fold: Rhomboid-like
Superfamily: Rhomboid-like
Family: Rhomboid-like
Phyre2
| 3 |
|
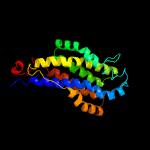
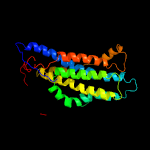
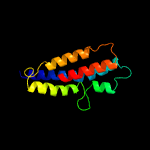
PDB 2gfp chain A
Region: 93 - 240
Aligned: 148
Modelled: 148
Confidence: 79.6%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
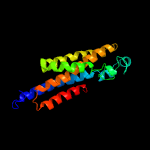
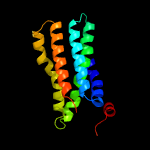
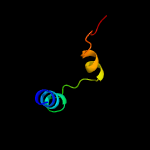
PDB 4zp0 chain A
Region: 30 - 235
Aligned: 195
Modelled: 206
Confidence: 65.4%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:multidrug transporter mdfa;
PDBTitle: crystal structure of e. coli multidrug transporter mdfa in complex2 with deoxycholate
Phyre2
| 5 |
|
PDB 6e9o chain A
Region: 30 - 234
Aligned: 194
Modelled: 205
Confidence: 55.5%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:d-galactonate transport;
PDBTitle: e. coli d-galactonate:proton symporter mutant e133q in the outward2 substrate-bound form
Phyre2
| 6 |
|
PDB 4iu8 chain A
Region: 22 - 192
Aligned: 160
Modelled: 170
Confidence: 29.2%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:nitrite extrusion protein 2;
PDBTitle: crystal structure of a membrane transporter (selenomethionine2 derivative)
Phyre2
| 7 |
|
PDB 3wdo chain A
Region: 28 - 237
Aligned: 199
Modelled: 210
Confidence: 28.0%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:mfs transporter;
PDBTitle: structure of e. coli yajr transporter
Phyre2
| 8 |
|
PDB 6h7d chain A
Region: 93 - 218
Aligned: 126
Modelled: 126
Confidence: 27.3%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:sugar transport protein 10;
PDBTitle: crystal structure of a. thaliana sugar transport protein 10 in complex2 with glucose in the outward occluded state
Phyre2
| 9 |
|
PDB 1pw4 chain A
Region: 18 - 239
Aligned: 214
Modelled: 222
Confidence: 23.5%
Identity: 9%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 10 |
|
PDB 6g9x chain B
Region: 93 - 234
Aligned: 142
Modelled: 142
Confidence: 19.7%
Identity: 12%
PDB header:membrane protein
Chain: B: PDB Molecule:major facilitator superfamily mfs_1;
PDBTitle: crystal structure of a mfs transporter at 2.54 angstroem resolution
Phyre2
| 11 |
|
PDB 5guf chain A
Region: 196 - 218
Aligned: 23
Modelled: 23
Confidence: 19.4%
Identity: 35%
PDB header:transferase
Chain: A: PDB Molecule:cdp-archaeol synthase;
PDBTitle: structural insight into an intramembrane enzyme for archaeal membrane2 lipids biosynthesis
Phyre2
| 12 |
|
PDB 4lds chain B
Region: 42 - 239
Aligned: 191
Modelled: 198
Confidence: 11.9%
Identity: 16%
PDB header:transport protein, membrane protein
Chain: B: PDB Molecule:bicyclomycin resistance protein tcab;
PDBTitle: the inward-facing structure of the glucose transporter from2 staphylococcus epidermidis
Phyre2
| 13 |
|
PDB 5xpd chain A
Region: 81 - 238
Aligned: 157
Modelled: 158
Confidence: 11.5%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sugar transporter;
PDBTitle: sugar transporter of atsweet13 in inward-facing state with a substrate2 analog
Phyre2
| 14 |
|
PDB 4j05 chain A
Region: 34 - 157
Aligned: 117
Modelled: 124
Confidence: 7.6%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:phosphate transporter;
PDBTitle: crystal structure of a eukaryotic phosphate transporter
Phyre2
| 15 |
|
PDB 2b6p chain A
Region: 205 - 239
Aligned: 35
Modelled: 35
Confidence: 5.7%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: x-ray structure of lens aquaporin-0 (aqp0) (lens mip) in an open pore2 state
Phyre2