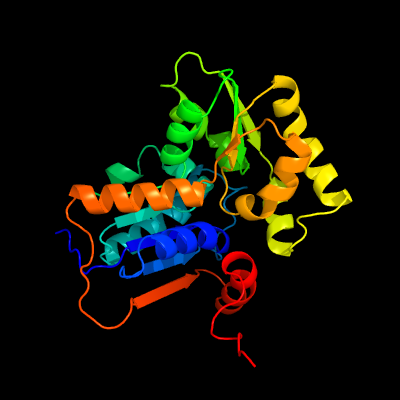
| 1 | c5ijwA_
|
|
 |
100.0 |
87 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase (muri) from mycobacterium smegmatis with bound d-2 glutamate, 1.8 angstrom resolution, x-ray diffraction
|
|
|
|
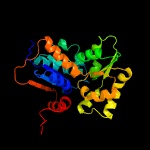
| 2 | c2jfoB_
|
|
 |
100.0 |
40 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
|
|
|
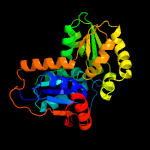
| 3 | c3hfrA_
|
|
 |
100.0 |
41 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
|
|
|
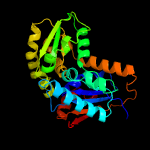
| 4 | c2dwuA_
|
|
 |
100.0 |
40 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
|
|
|
| 5 | c2gzmB_
|
|
 |
100.0 |
39 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus anthracis
|
|
|
|
| 6 | c2jfqA_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate racemase in2 complex with d- glutamate
|
|
|
|
| 7 | c2ohoA_
|
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
|
|
|
| 8 | c3outC_
|
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: C: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
|
|
|
|
| 9 | c1zuwA_
|
|
 |
100.0 |
42 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
|
|
|
| 10 | c5w16D_
|
|
 |
100.0 |
45 |
PDB header:isomerase
Chain: D: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from thermus thermophilus in2 complex with d-glutamate
|
|
|
|
| 11 | c2jfnA_
|
|
 |
100.0 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
|
|
|
|
| 12 | c3uhfB_
|
|
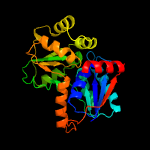 |
100.0 |
32 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from campylobacter jejuni2 subsp. jejuni
|
|
|
|
| 13 | c1b74A_
|
|
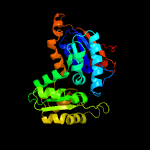 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
|
|
|
| 14 | c2jfzB_
|
|
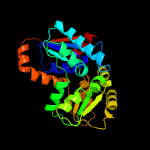 |
100.0 |
30 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|
|
|
|
| 15 | c5b19A_
|
|
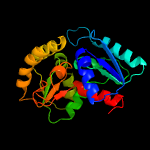 |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:aspartate racemase;
PDBTitle: picrophilus torridus aspartate racemase
|
|
|
|
| 16 | d1b74a1
|
|
 |
100.0 |
42 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
|
| 17 | c5wxzA_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:mcyf;
PDBTitle: crystal structure of microcystis aeruginosa pcc 7806 aspartate2 racemase in complex with d-aspartate
|
|
|
|
| 18 | c2zskA_
|
|
 |
100.0 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
|
|
|
| 19 | c5elmB_
|
|
 |
100.0 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:asp/glu_racemase family protein;
PDBTitle: crystal structure of l-aspartate/glutamate specific racemase in2 complex with l-glutamate
|
|
|
|
| 20 | c3s81A_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
|
|
|
| 21 | c3ojcD_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:putative aspartate/glutamate racemase;
PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
|
|
|
| 22 | c2dx7B_ |
|
not modelled |
100.0 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:aspartate racemase;
PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
|
|
|
| 23 | d1b74a2 |
|
not modelled |
100.0 |
31 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
| 24 | c3qvjB_ |
|
not modelled |
100.0 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
|
|
| 25 | c2vlbC_ |
|
not modelled |
99.9 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
|
|
| 26 | c5lg5F_ |
|
not modelled |
99.9 |
15 |
PDB header:isomerase
Chain: F: PDB Molecule:allantoin racemase;
PDBTitle: crystal structure of allantoin racemase from pseudomonas fluorescens2 allr
|
|
|
| 27 | c2eq5D_ |
|
not modelled |
99.9 |
20 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
|
|
| 28 | c2dgdD_ |
|
not modelled |
99.8 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
|
|
| 29 | c5b19B_ |
|
not modelled |
99.8 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:aspartate racemase;
PDBTitle: picrophilus torridus aspartate racemase
|
|
|
| 30 | d1jfla2 |
|
not modelled |
99.8 |
20 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
| 31 | d2dx7a1 |
|
not modelled |
99.7 |
18 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
| 32 | d1jfla1 |
|
not modelled |
99.3 |
21 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
| 33 | c4ix1B_ |
|
not modelled |
98.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of hypothetical protein opag_01669 from rhodococcus2 opacus pd630, target 016205
|
|
|
| 34 | c4fq5B_ |
|
not modelled |
98.5 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:maleate cis-trans isomerase;
PDBTitle: crystal structure of the maleate isomerase iso(c200a) from pseudomonas2 putida s16 with maleate
|
|
|
| 35 | c2xecD_ |
|
not modelled |
98.5 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
|
|
| 36 | c5tpwB_ |
|
not modelled |
94.5 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor ionotropic, nmda 2a;
PDBTitle: crystal structure of amino terminal domains of the nmda receptor2 subunit glun1 and glun2a in complex with zinc at the glun2a
|
|
|
| 37 | d1m3ua_ |
|
not modelled |
94.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 38 | c5uowB_ |
|
not modelled |
93.9 |
12 |
PDB header:membrane protein
Chain: B: PDB Molecule:n-methyl-d-aspartate receptor subunit nr2a;
PDBTitle: triheteromeric nmda receptor glun1/glun2a/glun2b in complex with2 glycine, glutamate, mk-801 and a glun2b-specific fab, at ph 6.5
|
|
|
| 39 | c5kc9B_ |
|
not modelled |
92.3 |
15 |
PDB header:signaling protein
Chain: B: PDB Molecule:glutamate receptor ionotropic, delta-1;
PDBTitle: crystal structure of the amino-terminal domain (atd) of iglur delta-12 (glud1)
|
|
|
| 40 | c3ez4B_ |
|
not modelled |
92.2 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate hydroxymethyltransferase2 from burkholderia pseudomallei
|
|
|
| 41 | c2wjxA_ |
|
not modelled |
91.6 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: crystal structure of the human ionotropic glutamate2 receptor glur2 atd region at 4.1 a resolution
|
|
|
| 42 | d1o66a_ |
|
not modelled |
91.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 43 | c2oblA_ |
|
not modelled |
90.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:escn;
PDBTitle: structural and biochemical analysis of a prototypical atpase from the2 type iii secretion system of pathogenic bacteria
|
|
|
| 44 | c2h31A_ |
|
not modelled |
90.0 |
14 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
|
|
| 45 | d1o5oa_ |
|
not modelled |
88.6 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 46 | c3gkaB_ |
|
not modelled |
88.6 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
|
|
| 47 | d1qcza_ |
|
not modelled |
88.6 |
15 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 48 | c4nphA_ |
|
not modelled |
88.3 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:probable secretion system apparatus atp synthase ssan;
PDBTitle: crystal structure of ssan from salmonella enterica
|
|
|
| 49 | c4ja0A_ |
|
not modelled |
88.2 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase;
PDBTitle: crystal structure of the invertebrate bi-functional purine2 biosynthesis enzyme paics at 2.8 a resolution
|
|
|
| 50 | c6njpC_ |
|
not modelled |
88.1 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:translocator escn;
PDBTitle: structure of the assembled atpase escn in complex with its central2 stalk esco from the enteropathogenic e. coli (epec) type iii3 secretion system
|
|
|
| 51 | c3p3wC_ |
|
not modelled |
88.0 |
16 |
PDB header:transport protein
Chain: C: PDB Molecule:glutamate receptor 3;
PDBTitle: structure of a dimeric glua3 n-terminal domain (ntd) at 4.2 a2 resolution
|
|
|
| 52 | c5ideA_ |
|
not modelled |
87.7 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: cryo-em structure of glua2/3 ampa receptor heterotetramer (model i)
|
|
|
| 53 | c3lp6D_ |
|
not modelled |
87.5 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
|
|
| 54 | c3kg2A_ |
|
not modelled |
87.5 |
13 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
|
|
|
| 55 | d1skyb3 |
|
not modelled |
87.0 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
|
|
| 56 | c6qkzA_ |
|
not modelled |
86.6 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:glua1;
PDBTitle: full length glua1/2-gamma8 complex
|
|
|
| 57 | c2fw9A_ |
|
not modelled |
86.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
|
|
| 58 | d1e5qa1 |
|
not modelled |
86.0 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 59 | d1u11a_ |
|
not modelled |
85.8 |
15 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 60 | c3orsD_ |
|
not modelled |
85.6 |
15 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
|
|
| 61 | c2dpyA_ |
|
not modelled |
85.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:flagellum-specific atp synthase;
PDBTitle: crystal structure of the flagellar type iii atpase flii
|
|
|
| 62 | c5bo5B_ |
|
not modelled |
84.2 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:neq263;
PDBTitle: structure of a unique atp synthase subunit neqb from nanoarcheaum2 equitans
|
|
|
| 63 | c5cjjA_ |
|
not modelled |
84.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
| 64 | c2ywrA_ |
|
not modelled |
84.1 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex aeolicus
|
|
|
| 65 | d1oy0a_ |
|
not modelled |
84.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 66 | c3tqrA_ |
|
not modelled |
83.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
|
|
| 67 | c5eoxB_ |
|
not modelled |
83.7 |
29 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:type 4 fimbrial biogenesis protein pilm;
PDBTitle: pseudomonas aeruginosa pilm bound to adp
|
|
|
| 68 | d1jkxa_ |
|
not modelled |
83.6 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 69 | c3h5lB_ |
|
not modelled |
83.0 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc transporter;
PDBTitle: crystal structure of a putative branched-chain amino acid abc2 transporter from silicibacter pomeroyi
|
|
|
| 70 | c6irfD_ |
|
not modelled |
82.9 |
10 |
PDB header:membrane protein
Chain: D: PDB Molecule:glutamate receptor ionotropic, nmda 2a;
PDBTitle: structure of the human glun1/glun2a nmda receptor in the2 glutamate/glycine-bound state at ph 6.3, class i
|
|
|
| 71 | c4ew6A_ |
|
not modelled |
82.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-galactose-1-dehydrogenase protein;
PDBTitle: crystal structure of d-galactose-1-dehydrogenase protein from2 rhizobium etli
|
|
|
| 72 | c4grdA_ |
|
not modelled |
82.3 |
21 |
PDB header:lyase,isomerase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from burkholderia cenocepacia j2315
|
|
|
| 73 | d1meoa_ |
|
not modelled |
82.0 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 74 | c5syrA_ |
|
not modelled |
81.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable atp synthase spal/mxib;
PDBTitle: crystal structure of atpase delta1-79 spa47 r350a
|
|
|
| 75 | c1dkrB_ |
|
not modelled |
81.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
|
|
| 76 | c3cpqB_ |
|
not modelled |
81.6 |
10 |
PDB header:ribosomal protein
Chain: B: PDB Molecule:50s ribosomal protein l30e;
PDBTitle: crystal structure of l30e a ribosomal protein from2 methanocaldococcus jannaschii dsm2661 (mj1044)
|
|
|
| 77 | d1xmpa_ |
|
not modelled |
80.9 |
21 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 78 | c3p9xB_ |
|
not modelled |
80.8 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
|
|
| 79 | c5dmnA_ |
|
not modelled |
80.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:homocysteine s-methyltransferase;
PDBTitle: crystal structure of the homocysteine methyltransferase mmum from2 escherichia coli, apo form
|
|
|
| 80 | c3trhI_ |
|
not modelled |
80.7 |
17 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
|
|
| 81 | c4b4kK_ |
|
not modelled |
80.6 |
21 |
PDB header:isomerase
Chain: K: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of bacillus anthracis pure
|
|
|
| 82 | c3bolB_ |
|
not modelled |
80.1 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
|
|
| 83 | d1i5ea_ |
|
not modelled |
79.9 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 84 | c3sg0A_ |
|
not modelled |
79.7 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
|
|
|
| 85 | c6o55B_ |
|
not modelled |
79.4 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 (pure) from legionella pneumophila
|
|
|
| 86 | d1w41a1 |
|
not modelled |
79.3 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 87 | c2c61A_ |
|
not modelled |
78.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:a-type atp synthase non-catalytic subunit b;
PDBTitle: crystal structure of the non-catalytic b subunit of a-type2 atpase from m. mazei go1
|
|
|
| 88 | c3av3A_ |
|
not modelled |
78.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 geobacillus kaustophilus
|
|
|
| 89 | d1lt7a_ |
|
not modelled |
78.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
|
|
| 90 | c5b0oB_ |
|
not modelled |
78.4 |
17 |
PDB header:hydrolase/motor protein
Chain: B: PDB Molecule:flagellum-specific atp synthase;
PDBTitle: structure of the flih-flii complex
|
|
|
| 91 | c3a5dM_ |
|
not modelled |
77.5 |
18 |
PDB header:hydrolase
Chain: M: PDB Molecule:v-type atp synthase beta chain;
PDBTitle: inter-subunit interaction and quaternary rearrangement2 defined by the central stalk of prokaryotic v1-atpase
|
|
|
| 92 | d2bo1a1 |
|
not modelled |
77.5 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 93 | c4s1nA_ |
|
not modelled |
77.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
|
|
|
| 94 | d1umya_ |
|
not modelled |
77.3 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
|
|
| 95 | c1u9yD_ |
|
not modelled |
77.2 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
|
|
| 96 | d1o4va_ |
|
not modelled |
77.1 |
14 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 97 | c6focB_ |
|
not modelled |
77.0 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:atp synthase subunit alpha,atp synthase subunit alpha,atp
PDBTitle: f1-atpase from mycobacterium smegmatis
|
|
|
| 98 | c4mwaA_ |
|
not modelled |
76.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
| 99 | c5ereA_ |
|
not modelled |
75.9 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: extracellular ligand binding receptor from desulfohalobium retbaense2 dsm5692
|
|
|
| 100 | c4a3uB_ |
|
not modelled |
74.8 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 101 | c3lwzC_ |
|
not modelled |
74.4 |
37 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-dehydroquinate2 dehydratase (aroq) from yersinia pestis
|
|
|
| 102 | d1t0kb_ |
|
not modelled |
74.0 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 103 | d1skye3 |
|
not modelled |
73.9 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
|
|
| 104 | c2gr2A_ |
|
not modelled |
73.9 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
|
|
| 105 | c2pjuD_ |
|
not modelled |
73.5 |
17 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon regulatory protein2 prpr
|
|
|
| 106 | d1uqra_ |
|
not modelled |
73.4 |
26 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 107 | c3zu0A_ |
|
not modelled |
72.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
|
|
| 108 | c6re7U_ |
|
not modelled |
72.5 |
19 |
PDB header:proton transport
Chain: U: PDB Molecule:atp synthase subunit alpha;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 2c,2 focussed refinement of f1 head and rotor
|
|
|
| 109 | c1nhqA_ |
|
not modelled |
72.4 |
20 |
PDB header:oxidoreductase (h2o2(a))
Chain: A: PDB Molecule:nadh peroxidase;
PDBTitle: crystallographic analyses of nadh peroxidase cys42ala and cys42ser2 mutants: active site structure, mechanistic implications, and an3 unusual environment of arg303
|
|
|
| 110 | d1gqoa_ |
|
not modelled |
72.1 |
31 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 111 | c3lpnB_ |
|
not modelled |
72.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
|
|
|
| 112 | c2o14A_ |
|
not modelled |
71.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yxim;
PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus subtilis.2 northeast structural genomics consortium target sr595
|
|
|
| 113 | c2v3cC_ |
|
not modelled |
71.8 |
18 |
PDB header:signaling protein
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
|
|
|
| 114 | c4a1dG_ |
|
not modelled |
71.7 |
11 |
PDB header:ribosome
Chain: G: PDB Molecule:rpl30;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
|
|
| 115 | d2cvza2 |
|
not modelled |
71.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 116 | c3j21Z_ |
|
not modelled |
71.1 |
13 |
PDB header:ribosome
Chain: Z: PDB Molecule:50s ribosomal protein l30e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
|
|
|
| 117 | c5ocsB_ |
|
not modelled |
71.0 |
20 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 118 | c3wb9A_ |
|
not modelled |
70.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diaminopimelate dehydrogenase;
PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
|
|
|
| 119 | c3cr8C_ |
|
not modelled |
70.3 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:sulfate adenylyltransferase, adenylylsulfate kinase;
PDBTitle: hexameric aps kinase from thiobacillus denitrificans
|
|
|
| 120 | c2qe7C_ |
|
not modelled |
70.1 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:atp synthase subunit alpha;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
|
|
|