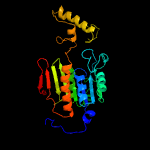
1 c2ymvA_
100.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: acg nitroreductase;PDBTitle: structure of reduced m smegmatis 5246, a homologue of m.2 tuberculosis acg
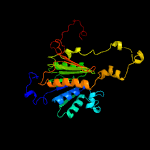
2 c6h77B_
100.0
17
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme 5;PDBTitle: e1 enzyme for ubiquitin like protein activation in complex with ubl
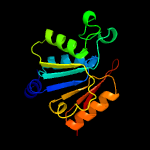
3 c4d7aA_
100.0
19
PDB header: ligaseChain: A: PDB Molecule: trna threonylcarbamoyladenosine dehydratase;PDBTitle: crystal structure of e. coli trna n6-threonylcarbamoyladenosine2 dehydratase, tcda, in complex with amp at 1.801 angstroem3 resolution
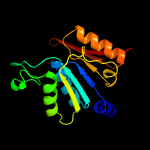
4 c1zfnA_
100.0
21
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase thif;PDBTitle: structural analysis of escherichia coli thif
5 c3h9gA_
100.0
18
PDB header: transferase/antibioticChain: A: PDB Molecule: mccb protein;PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
6 d1jw9b_
100.0
24
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Molybdenum cofactor biosynthesis protein MoeB
7 c5ff5A_
100.0
26
PDB header: transferaseChain: A: PDB Molecule: paaa;PDBTitle: crystal structure of semet paaa
8 d1yovb1
100.0
21
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)
9 c3gznB_
100.0
16
PDB header: protein binding/ligaseChain: B: PDB Molecule: nedd8-activating enzyme e1 catalytic subunit;PDBTitle: structure of nedd8-activating enzyme in complex with nedd8 and mln4924
10 c3kycB_
100.0
20
PDB header: ligaseChain: B: PDB Molecule: sumo-activating enzyme subunit 2;PDBTitle: human sumo e1 complex with a sumo1-amp mimic
11 c3kydB_
100.0
20
PDB header: ligaseChain: B: PDB Molecule: sumo-activating enzyme subunit 2;PDBTitle: human sumo e1~sumo1-amp tetrahedral intermediate mimic
12 c1y8qD_
100.0
20
PDB header: ligaseChain: D: PDB Molecule: ubiquitin-like 2 activating enzyme e1b;PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
13 c3wv9D_
100.0
16
PDB header: transferaseChain: D: PDB Molecule: hmd co-occurring protein hcge;PDBTitle: guanylylpyridinol (gp)- and atp-bound hcge from methanothermobacter2 marburgensis
14 c6dc6A_
100.0
21
PDB header: signaling protein/ligaseChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme 1;PDBTitle: crystal structure of human ubiquitin activating enzyme e1 (uba1) in2 complex with ubiquitin
15 c3vh3A_
100.0
14
PDB header: metal binding protein/protein transportChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of atg7ctd-atg8 complex
16 c4ii3A_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-activating enzyme e1 1;PDBTitle: crystal structure of s. pombe ubiquitin activating enzyme 1 (uba1) in2 complex with ubiquitin and atp/mg
17 c3cmmA_
100.0
21
PDB header: ligase/protein bindingChain: A: PDB Molecule: ubiquitin-activating enzyme e1 1;PDBTitle: crystal structure of the uba1-ubiquitin complex
18 c1y8qA_
100.0
16
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-like 1 activating enzyme e1a;PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
19 c2nvuB_
100.0
16
PDB header: protein turnover, ligaseChain: B: PDB Molecule: maltose binding protein/nedd8-activating enzymePDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
20 c3vh1A_
100.0
17
PDB header: metal binding proteinChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
21 d1yova1
not modelled
100.0
16
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)
22 c4p22A_
not modelled
100.0
23
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme 1;PDBTitle: crystal structure of n-terminal fragments of e1
23 c3gucB_
not modelled
99.9
14
PDB header: transferaseChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme 5;PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
24 c3gr3B_
not modelled
99.7
13
PDB header: flavoproteinChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a nitroreductase-like family protein (pnba,2 bh06130) from bartonella henselae str. houston-1 at 1.45 a resolution
25 c2islB_
not modelled
99.5
19
PDB header: flavoproteinChain: B: PDB Molecule: blub;PDBTitle: blub bound to reduced flavin (fmnh2) and molecular oxygen.2 (clear crystal form)
26 c3kwkA_
not modelled
99.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh dehydrogenase/nad(p)h nitroreductase;PDBTitle: crystal structure of putative nadh dehydrogenase/nad(p)h2 nitroreductase (np_809094.1) from bacteroides thetaiotaomicron vpi-3 5482 at 1.54 a resolution
27 c3gh8A_
not modelled
99.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: iodotyrosine dehalogenase 1;PDBTitle: crystal structure of mus musculus iodotyrosine deiodinase (iyd) bound2 to fmn and di-iodotyrosine (dit)
28 c4xomB_
not modelled
99.5
19
PDB header: unknown functionChain: B: PDB Molecule: coenzyme f420:l-glutamate ligase;PDBTitle: coenzyme f420:l-glutamate ligase (fbib) from mycobacterium2 tuberculosis (c-terminal domain).
29 c3eo8A_
not modelled
99.5
13
PDB header: flavoproteinChain: A: PDB Molecule: blub-like flavoprotein;PDBTitle: crystal structure of blub-like flavoprotein (yp_001089088.1) from2 clostridium difficile 630 at 1.74 a resolution
30 c3to0A_
not modelled
99.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: iodotyrosine deiodinase 1;PDBTitle: crystal structure of mus musculus iodotyrosine deiodinase (iyd) c217a,2 c239a bound to fmn
31 c3e39A_
not modelled
99.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (dde_0787) from desulfovibrio desulfuricans subsp. at 1.70 a3 resolution
32 c2i7hE_
not modelled
99.4
18
PDB header: oxidoreductaseChain: E: PDB Molecule: nitroreductase-like family protein;PDBTitle: crystal structure of the nitroreductase-like family protein from2 bacillus cereus
33 c2wzvB_
not modelled
99.4
19
PDB header: oxidoreductaseChain: B: PDB Molecule: nfnb protein;PDBTitle: crystal structure of the fmn-dependent nitroreductase nfnb2 from mycobacterium smegmatis
34 c5ko8B_
not modelled
99.4
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of haliscomenobacter hydrossis iodotyrosine2 deiodinase (iyd) bound to fmn and mono-iodotyrosine (i-tyr)
35 c3ek3A_
not modelled
99.4
15
PDB header: flavoproteinChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of nitroreductase with bound fmn (yp_211706.1) from2 bacteroides fragilis nctc 9343 at 1.70 a resolution
36 d1noxa_
not modelled
99.3
15
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
37 c3pxvD_
not modelled
99.3
17
PDB header: oxidoreductaseChain: D: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a nitroreductase with bound fmn (dhaf_2018) from2 desulfitobacterium hafniense dcb-2 at 2.30 a resolution
38 c3bm2B_
not modelled
99.3
12
PDB header: oxidoreductaseChain: B: PDB Molecule: protein ydja;PDBTitle: crystal structure of a minimal nitroreductase ydja from escherichia2 coli k12 with and without fmn cofactor
39 c3m5kA_
not modelled
99.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh dehydrogenase/nad(p)h nitroreductase;PDBTitle: crystal structure of putative nadh dehydrogenase/nad(p)h2 nitroreductase (bdi_1728) from parabacteroides distasonis atcc 85033 at 1.86 a resolution
40 d1bkja_
not modelled
99.3
18
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
41 d1vfra_
not modelled
99.3
13
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
42 d1kqba_
not modelled
99.3
16
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
43 d1f5va_
not modelled
99.3
18
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
44 c3n2sD_
not modelled
99.3
19
PDB header: oxidoreductaseChain: D: PDB Molecule: nadph-dependent nitro/flavin reductase;PDBTitle: structure of nfra1 nitroreductase from b. subtilis
45 c3g14B_
not modelled
99.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of nitroreductase family protein (yp_877874.1) from2 clostridium novyi nt at 1.75 a resolution
46 d1zcha1
not modelled
99.2
16
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
47 c4eo3A_
not modelled
99.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein/nadh dehydrogenase;PDBTitle: peroxiredoxin nitroreductase fusion enzyme
48 d1ykia1
not modelled
99.2
11
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
49 c5hdjA_
not modelled
99.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nfra1;PDBTitle: structure of b. megaterium nfra1
50 c3gfaB_
not modelled
99.2
13
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (cd3205) from clostridium difficile 630 at 1.35 a resolution
51 c5heiE_
not modelled
99.2
20
PDB header: oxidoreductaseChain: E: PDB Molecule: nfra2;PDBTitle: structure of b. megaterium nfra2
52 d2b67a1
not modelled
99.2
12
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
53 c3bemA_
not modelled
99.2
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nad(p)h nitroreductase ydfn;PDBTitle: crystal structure of putative nitroreductase ydfn (2632848) from2 bacillus subtilis at 1.65 a resolution
54 c3e10B_
not modelled
99.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadh oxidase;PDBTitle: crystal structure of putative nadh oxidase (np_348178.1) from2 clostridium acetobutylicum at 1.40 a resolution
55 c3ge5A_
not modelled
99.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nad(p)h:fmn oxidoreductase;PDBTitle: crystal structure of a putative nad(p)h:fmn oxidoreductase (pg0310)2 from porphyromonas gingivalis w83 at 1.70 a resolution
56 c4qlyB_
not modelled
99.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: enone reductase cla-er;PDBTitle: crystal structure of cla-er, a novel enone reductase catalyzing a key2 step of a gut-bacterial fatty acid saturation metabolism,3 biohydrogenation
57 c4dn2A_
not modelled
99.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of putative nitroreductase from geobacter2 metallireducens gs-15
58 c3k6hB_
not modelled
99.1
13
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a nitroreductase family protein from2 agrobacterium tumefaciens str. c58
59 c3koqC_
not modelled
99.1
13
PDB header: oxidoreductaseChain: C: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a nitroreductase family protein (cd3355) from2 clostridium difficile 630 at 1.58 a resolution
60 c5j6cA_
not modelled
99.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase;PDBTitle: fmn-dependent nitroreductase (cdr20291_0767) from clostridium2 difficile r20291
61 c6czpH_
not modelled
99.1
12
PDB header: oxidoreductaseChain: H: PDB Molecule: oxygen-insensitive nad(p)h nitroreductase;PDBTitle: 2.2 angstrom resolution crystal structure oxygen-insensitive nad(p)h-2 dependent nitroreductase nfsb from vibrio vulnificus in complex with3 fmn
62 c3of4A_
not modelled
99.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a fmn/fad- and nad(p)h-dependent nitroreductase2 (nfnb, il2077) from idiomarina loihiensis l2tr at 1.90 a resolution
63 c3eofB_
not modelled
99.1
12
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase (yp_213212.1) from2 bacteroides fragilis nctc 9343 at 1.99 a resolution
64 c3qdlD_
not modelled
99.1
17
PDB header: oxidoreductaseChain: D: PDB Molecule: oxygen-insensitive nadph nitroreductase;PDBTitle: crystal structure of rdxa from helicobacter pyroli
65 c3gagB_
not modelled
99.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadh dehydrogenase, nadph nitroreductase;PDBTitle: crystal structure of a nitroreductase-like protein (smu.346) from2 streptococcus mutans at 1.70 a resolution
66 d2frea1
not modelled
99.0
23
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
67 c3ge6B_
not modelled
99.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (exig_2970) from exiguobacterium sibiricum 255-15 at 1.85 a3 resolution
68 c3gbhC_
not modelled
99.0
10
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p)h-flavin oxidoreductase;PDBTitle: crystal structure of a putative nad(p)h:fmn oxidoreductase (se1966)2 from staphylococcus epidermidis atcc 12228 at 2.00 a resolution
69 c2r01A_
not modelled
99.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a putative fmn-dependent nitroreductase (ct0345)2 from chlorobium tepidum tls at 1.15 a resolution
70 c2hayD_
not modelled
99.0
14
PDB header: oxidoreductaseChain: D: PDB Molecule: putative nad(p)h-flavin oxidoreductase;PDBTitle: the crystal structure of the putative nad(p)h-flavin oxidoreductase2 from streptococcus pyogenes m1 gas
71 c2wqfA_
not modelled
98.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: copper induced nitroreductase d;PDBTitle: crystal structure of the nitroreductase cind from2 lactococcus lactis in complex with fmn
72 c2h0uA_
not modelled
98.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph-flavin oxidoreductase;PDBTitle: crystal structure of nad(p)h-flavin oxidoreductase from helicobacter2 pylori
73 d2ifaa1
not modelled
98.8
15
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
74 c3hoiA_
not modelled
98.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nitroreductase bf3017;PDBTitle: crystal structure of fmn-dependent nitroreductase bf3017 from2 bacteroides fragilis nctc 9343 (yp_212631.1) from bacteroides3 fragilis nctc 9343 at 1.55 a resolution
75 d1ywqa1
not modelled
98.7
16
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
76 c4urpB_
not modelled
98.7
14
PDB header: oxidoreductaseChain: B: PDB Molecule: fatty acid repression mutant protein 2;PDBTitle: the crystal structure of nitroreductase from saccharomyces2 cerevisiae
77 c5j62B_
not modelled
98.7
11
PDB header: oxidoreductaseChain: B: PDB Molecule: putative reductase;PDBTitle: fmn-dependent nitroreductase (cdr20291_0684) from clostridium2 difficile r20291
78 c3hj9A_
not modelled
98.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of a putative nitroreductase (reut_a1228) from2 ralstonia eutropha jmp134 at 2.00 a resolution
79 c3eo7A_
not modelled
98.6
18
PDB header: flavoproteinChain: A: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase (ava_2154) from2 anabaena variabilis atcc 29413 at 1.80 a resolution
80 d1vkwa_
not modelled
98.5
25
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: Putative nitroreductase TM1586
81 c5lq4B_
not modelled
98.2
16
PDB header: oxidoreductaseChain: B: PDB Molecule: cyagox;PDBTitle: the structure of thcox, the first oxidase protein from the cyanobactin2 pathways
82 c2axqA_
not modelled
98.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
83 c1e5lA_
not modelled
97.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine reductase;PDBTitle: apo saccharopine reductase from magnaporthe grisea
84 c4rl6A_
not modelled
97.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q04l03_strp2 protein from streptococcus2 pneumoniae. northeast structural genomics consortium target spr105
85 c5l78A_
not modelled
97.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-aminoadipic semialdehyde synthase, mitochondrial;PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
86 d1pjqa1
not modelled
97.4
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like
87 c2z2vA_
not modelled
97.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
88 c3ic5A_
not modelled
96.8
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
89 d1vi2a1
not modelled
96.7
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
90 c4plpB_
not modelled
96.7
18
PDB header: transferaseChain: B: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase (hss) from2 blastochloris viridis in complex with nad
91 c5ugjC_
not modelled
96.6
17
PDB header: oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
92 c2d3tB_
not modelled
96.6
16
PDB header: lyase, oxidoreductase/transferaseChain: B: PDB Molecule: fatty oxidation complex alpha subunit;PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
93 c2nloA_
not modelled
96.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
94 d1e5qa1
not modelled
96.6
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
95 c4ywjB_
not modelled
96.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate reductase (htpa2 reductase) from pseudomonas aeruginosa
96 c1vi2B_
not modelled
96.5
11
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase 2;PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
97 c1pjtB_
not modelled
96.5
20
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
98 c1drwA_
not modelled
96.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
99 c3tozA_
not modelled
96.5
7
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
100 c3mogA_
not modelled
96.4
22
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of 3-hydroxybutyryl-coa dehydrogenase from2 escherichia coli k12 substr. mg1655
101 c6iaqA_
not modelled
96.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase n-terminus domain-containingPDBTitle: structure of amine dehydrogenase from mycobacterium smegmatis
102 c4inaA_
not modelled
96.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q7mss8_wolsu protein from wolinella2 succinogenes. northeast structural genomics consortium target wsr35
103 c3fwnB_
not modelled
96.3
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
104 c2dc1A_
not modelled
96.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
105 c3k6jA_
not modelled
96.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: protein f01g10.3, confirmed by transcript evidence;PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
106 c4f3yA_
not modelled
96.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: x-ray crystal structure of dihydrodipicolinate reductase from2 burkholderia thailandensis
107 c2hjrK_
not modelled
96.1
18
PDB header: oxidoreductaseChain: K: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
108 c5tenH_
not modelled
96.1
13
PDB header: oxidoreductaseChain: H: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
109 c2x58B_
not modelled
96.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: the crystal structure of mfe1 liganded with coa
110 c1u4sA_
not modelled
96.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
111 c6hrdD_
not modelled
96.0
16
PDB header: oxidoreductaseChain: D: PDB Molecule: 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of m. tuberculosis fadb2 (rv0468)
112 c4om8B_
not modelled
95.9
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic2 acid (fhmpc) 5-dehydrogenase, an nad+ dependent dismutase.
113 c4ei8A_
not modelled
95.9
15
PDB header: replicationChain: A: PDB Molecule: plasmid replication protein repx;PDBTitle: crystal structure of bacillus cereus tubz, apo-form
114 c4bgvB_
not modelled
95.8
26
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: 1.8 a resolution structure of the malate dehydrogenase from2 picrophilus torridus in its apo form
115 c6iauB_
not modelled
95.8
19
PDB header: oxidoreductaseChain: B: PDB Molecule: amine dehydrogenase;PDBTitle: amine dehydrogenase from cystobacter fuscus in complex with nadp+ and2 cyclohexylamine
116 c1ldbA_
not modelled
95.8
18
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: apo-l-lactate dehydrogenase;PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
117 c3e18A_
not modelled
95.8
24
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of nad-binding protein from listeria innocua
118 c1zcjA_
not modelled
95.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase
119 c4e21B_
not modelled
95.8
17
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase (decarboxylating);PDBTitle: the crystal structure of 6-phosphogluconate dehydrogenase from2 geobacter metallireducens
120 c1pzfD_
not modelled
95.7
28
PDB header: oxidoreductaseChain: D: PDB Molecule: lactate dehydrogenase;PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate