| 1 | d1ezwa_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 2 | c1z69D_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
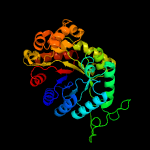
| 3 | d1f07a_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
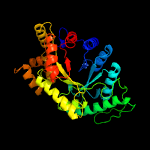
| 4 | c5tlcA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
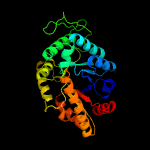
| 5 | d1luca_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
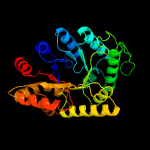
| 6 | d1lucb_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 7 | c3c8nB_
|
|
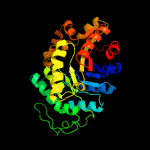 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 8 | c3sdoB_
|
|
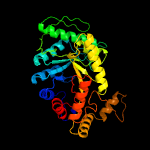 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
| 9 | c2wgkA_
|
|
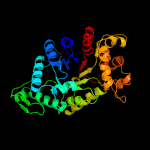 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
| 10 | c2i7gA_
|
|
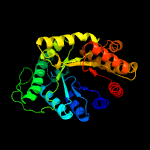 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
| 11 | d1rhca_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 12 | c5w4zA_
|
|
 |
100.0 |
16 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
| 13 | c1tvlA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 14 | d1tvla_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 15 | c3b9nB_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 16 | d1nqka_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 17 | c3raoB_
|
|
 |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
| 18 | c6friD_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
| 19 | c6ak1B_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
|
| 20 | c5dqpA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 21 | c5wanA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
| 22 | c2b81D_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
| 23 | d1nfpa_ |
|
not modelled |
99.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
99.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | c3qy6A_ |
|
not modelled |
68.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 26 | d1euca1 |
|
not modelled |
67.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 27 | c5zmyF_ |
|
not modelled |
67.0 |
15 |
PDB header:hydrolase
Chain: F: PDB Molecule:cis-epoxysuccinate hydrolase;
PDBTitle: crystal structure of a cis-epoxysuccinate hydrolase producing d(-)-2 tartaric acids
|
|
|
| 28 | d1o5ka_ |
|
not modelled |
53.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 29 | c1zfjA_ |
|
not modelled |
49.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
|
|
| 30 | d1v6ta_ |
|
not modelled |
48.0 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 31 | c2oztA_ |
|
not modelled |
38.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tlr1174 protein;
PDBTitle: crystal structure of o-succinylbenzoate synthase from2 thermosynechococcus elongatus bp-1
|
|
|
| 32 | c4b5nA_ |
|
not modelled |
37.3 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: crystal structure of oxidized shewanella yellow enzyme 4 (sye4)
|
|
|
| 33 | d1oyaa_ |
|
not modelled |
37.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 34 | c2ekcA_ |
|
not modelled |
35.6 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 35 | c4ggbA_ |
|
not modelled |
34.8 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a proposed galactarolactone cycloisomerase from2 agrobacterium tumefaciens, target efi-500704, with bound ca,3 disordered loops
|
|
|
| 36 | c1jvnB_ |
|
not modelled |
34.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
|
|
| 37 | c4nq1B_ |
|
not modelled |
34.3 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 38 | c2y7eA_ |
|
not modelled |
32.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
|
|
| 39 | d2d69a1 |
|
not modelled |
32.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 40 | c2gq8A_ |
|
not modelled |
32.6 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
|
|
| 41 | c4tmcB_ |
|
not modelled |
32.4 |
28 |
PDB header:flavoprotein
Chain: B: PDB Molecule:old yellow enzyme;
PDBTitle: crystal structure of old yellow enzyme from candida macedoniensis2 aku4588 complexed with p-hydroxybenzaldehyde
|
|
|
| 42 | c4m0xB_ |
|
not modelled |
32.2 |
6 |
PDB header:isomerase
Chain: B: PDB Molecule:chloromuconate cycloisomerase;
PDBTitle: crystal structure of 2-chloromuconate cycloisomerase from rhodococcus2 opacus 1cp
|
|
|
| 43 | c2wjeA_ |
|
not modelled |
32.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
|
|
| 44 | c4e8gA_ |
|
not modelled |
31.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme, n-terminal
PDBTitle: crystal structure of an enolase (mandelate racemase subgroup) from2 paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg
|
|
|
| 45 | d1q45a_ |
|
not modelled |
31.3 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 46 | c5n2pA_ |
|
not modelled |
31.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 47 | c1bf2A_ |
|
not modelled |
31.2 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: structure of pseudomonas isoamylase
|
|
|
| 48 | d1jdfa1 |
|
not modelled |
31.0 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 49 | c5dxxA_ |
|
not modelled |
29.8 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:artemisinic aldehyde delta(11(13)) reductase;
PDBTitle: crystal structure of dbr2
|
|
|
| 50 | d1rvka1 |
|
not modelled |
29.6 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 51 | c4exqA_ |
|
not modelled |
29.5 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase (upd) from2 burkholderia thailandensis e264
|
|
|
| 52 | c4a3uB_ |
|
not modelled |
29.4 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 53 | d1r3sa_ |
|
not modelled |
29.2 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
| 54 | c2d69B_ |
|
not modelled |
28.7 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
|
|
| 55 | c3fluD_ |
|
not modelled |
28.6 |
11 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
|
|
| 56 | d1z41a1 |
|
not modelled |
28.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 57 | c2yb1A_ |
|
not modelled |
28.4 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
|
|
|
| 58 | c3d0cB_ |
|
not modelled |
28.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 59 | c2h90A_ |
|
not modelled |
28.1 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
|
|
| 60 | d1m53a2 |
|
not modelled |
27.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 61 | d1uoka2 |
|
not modelled |
27.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 62 | d1vjia_ |
|
not modelled |
27.7 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 63 | c2qddA_ |
|
not modelled |
27.6 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a member of enolase superfamily from roseovarius2 nubinhibens ism
|
|
|
| 64 | d1vyra_ |
|
not modelled |
27.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 65 | d1lwha2 |
|
not modelled |
27.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 66 | d2dfaa1 |
|
not modelled |
27.3 |
11 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 67 | c3fkkA_ |
|
not modelled |
27.1 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 68 | c3eb2A_ |
|
not modelled |
26.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
|
|
| 69 | c4kemB_ |
|
not modelled |
26.9 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a tartrate dehydratase from azospirillum, target2 efi-502395, with bound mg and a putative acrylate ion, ordered active3 site
|
|
|
| 70 | c5zxgB_ |
|
not modelled |
26.5 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:cyclic maltosyl-maltose hydrolase;
PDBTitle: cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from arthrobacter2 globiformis, ligand-free form
|
|
|
| 71 | d2gdqa1 |
|
not modelled |
26.5 |
2 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 72 | c5m99A_ |
|
not modelled |
26.3 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: functional characterization and crystal structure of thermostable2 amylase from thermotoga petrophila, reveals high thermostability and3 an archaic form of dimerization
|
|
|
| 73 | c2r8wB_ |
|
not modelled |
26.2 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
|
|
| 74 | c6mqhA_ |
|
not modelled |
26.2 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate synthase (htpa2 synthase) from burkholderia mallei
|
|
|
| 75 | d1gwja_ |
|
not modelled |
26.1 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 76 | c3lyeA_ |
|
not modelled |
26.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 77 | c4fxsA_ |
|
not modelled |
26.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 78 | d1icpa_ |
|
not modelled |
25.8 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 79 | c2rfgB_ |
|
not modelled |
25.8 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
|
|
| 80 | c5kinC_ |
|
not modelled |
25.4 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 81 | c1ehaA_ |
|
not modelled |
25.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyltrehalose trehalohydrolase;
PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase from2 sulfolobus solfataricus
|
|
|
| 82 | c3fa4D_ |
|
not modelled |
25.3 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 83 | c3msyC_ |
|
not modelled |
25.1 |
22 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from a marine actinobacterium
|
|
|
| 84 | c2qdeA_ |
|
not modelled |
24.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing family2 protein from azoarcus sp. ebn1
|
|
|
| 85 | c4n4qD_ |
|
not modelled |
24.9 |
7 |
PDB header:lyase
Chain: D: PDB Molecule:acylneuraminate lyase;
PDBTitle: crystal structure of n-acetylneuraminate lyase from mycoplasma2 synoviae, crystal form ii
|
|
|
| 86 | c2hmcA_ |
|
not modelled |
24.6 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 87 | c2nuxB_ |
|
not modelled |
24.4 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
|
|
| 88 | c2qygC_ |
|
not modelled |
24.1 |
14 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
|
|
| 89 | c3m07A_ |
|
not modelled |
24.0 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:putative alpha amylase;
PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha amylase2 from salmonella typhimurium.
|
|
|
| 90 | c2r94B_ |
|
not modelled |
24.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
|
|
| 91 | c3si9B_ |
|
not modelled |
23.9 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
|
|
| 92 | d1nbwb_ |
|
not modelled |
23.8 |
17 |
Fold:Anticodon-binding domain-like
Superfamily:B12-dependent dehydatase associated subunit
Family:Dehydratase-reactivating factor beta subunit |
|
|
| 93 | d1oi7a1 |
|
not modelled |
23.7 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 94 | c3chvA_ |
|
not modelled |
23.6 |
5 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
|
|
| 95 | c2ya0A_ |
|
not modelled |
23.5 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading pneumococcal2 virulence factor spua
|
|
|
| 96 | d1hl2a_ |
|
not modelled |
23.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 97 | c3l5aA_ |
|
not modelled |
23.5 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
|
|
| 98 | c4xkyC_ |
|
not modelled |
23.2 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
|
|
|
| 99 | d1a3xa2 |
|
not modelled |
23.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 100 | d1xxxa1 |
|
not modelled |
23.2 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 101 | c4zr8B_ |
|
not modelled |
23.2 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: structure of uroporphyrinogen decarboxylase from acinetobacter2 baumannii
|
|
|
| 102 | c2ze0A_ |
|
not modelled |
23.1 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: alpha-glucosidase gsj
|
|
|
| 103 | d1wzla3 |
|
not modelled |
22.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 104 | c3c6cA_ |
|
not modelled |
22.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
|
|
| 105 | c3qfwB_ |
|
not modelled |
22.7 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
|
|
| 106 | c2ejaB_ |
|
not modelled |
22.6 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
|
|
| 107 | c2ehhE_ |
|
not modelled |
22.4 |
10 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
|
|
| 108 | c3bh1A_ |
|
not modelled |
22.4 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
|
|
| 109 | d1liua2 |
|
not modelled |
22.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 110 | c4icnB_ |
|
not modelled |
22.3 |
5 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from shewanella benthica
|
|
|
| 111 | c3noeA_ |
|
not modelled |
22.3 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
|
|
| 112 | c5ktlA_ |
|
not modelled |
22.2 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 113 | d1ea9c3 |
|
not modelled |
22.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 114 | d1gvia3 |
|
not modelled |
22.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 115 | c2pmqA_ |
|
not modelled |
22.2 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a mandelate racemase/muconate lactonizing enzyme2 from roseovarius sp. htcc2601
|
|
|
| 116 | c3b8iF_ |
|
not modelled |
22.1 |
11 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 117 | c2aaaA_ |
|
not modelled |
21.8 |
12 |
PDB header:glycosidase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction study at 2.12 angstroms resolution of two enzymes from aspergillus
|
|
|
| 118 | c2yxgD_ |
|
not modelled |
21.7 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
|
|
| 119 | c3gkaB_ |
|
not modelled |
21.6 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
|
|
| 120 | c2ze3A_ |
|
not modelled |
21.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|