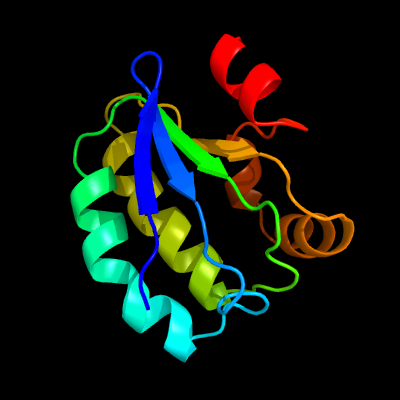
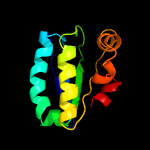
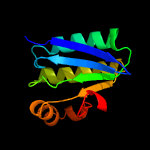
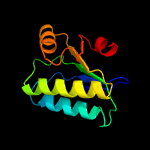
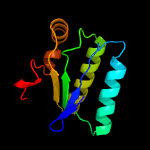
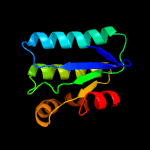
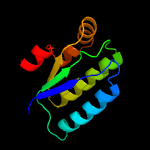
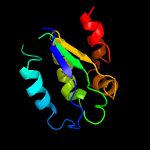
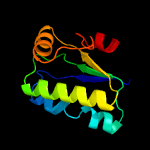
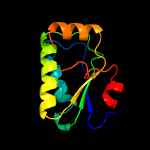
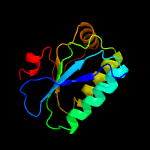
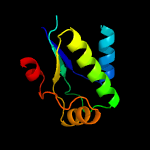
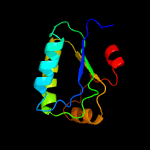
1 c3t6oA_
99.9
18
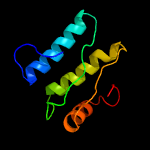
PDB header: transport proteinChain: A: PDB Molecule: sulfate transporter/antisigma-factor antagonist stas;PDBTitle: the structure of an anti-sigma-factor antagonist (stas) domain protein2 from planctomyces limnophilus.
2 c4hylB_
99.9
19
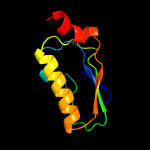
PDB header: transcription regulatorChain: B: PDB Molecule: stage ii sporulation protein;PDBTitle: the crystal structure of an anti-sigma-factor antagonist from2 haliangium ochraceum dsm 14365
3 d1th8b_
99.9
22
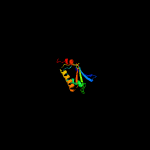
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa
4 d1auza_
99.9
21
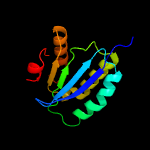
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa
5 d1vc1a_
99.9
23
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa
6 c4xs5D_
99.9
17
PDB header: transport proteinChain: D: PDB Molecule: sulfate transporter/antisigma-factor antagonist stas;PDBTitle: crystal structure of sulfate transporter/antisigma-factor antagonist2 stas from dyadobacter fermentans dsm 18053
7 c3f43A_
99.9
16
PDB header: transcriptionChain: A: PDB Molecule: putative anti-sigma factor antagonist tm1081;PDBTitle: crystal structure of a putative anti-sigma factor antagonist (tm1081)2 from thermotoga maritima at 1.59 a resolution
8 d1h4xa_
99.9
17
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Anti-sigma factor antagonist SpoIIaa
9 c2vy9A_
99.8
14
PDB header: gene regulationChain: A: PDB Molecule: anti-sigma-factor antagonist;PDBTitle: molecular architecture of the stressosome, a signal2 integration and transduction hub
10 c3mglA_
99.8
20
PDB header: transport proteinChain: A: PDB Molecule: sulfate permease family protein;PDBTitle: crystal structure of permease family protein from vibrio cholerae
11 c2klnA_
99.8
16
PDB header: transport proteinChain: A: PDB Molecule: probable sulphate-transport transmembrane protein, cog0659;PDBTitle: solution structure of stas domain of rv1739c from m. tuberculosis
12 c3oirA_
99.8
14
PDB header: transport proteinChain: A: PDB Molecule: sulfate transporter sulfate transporter family protein;PDBTitle: crystal structure of sulfate transporter family protein from wolinella2 succinogenes
13 c5ezbB_
99.8
15
PDB header: transport proteinChain: B: PDB Molecule: chicken prestin stas domain,chicken prestin stas domain;PDBTitle: chicken prestin stas domain
14 c3ny7A_
99.8
9
PDB header: membrane proteinChain: A: PDB Molecule: sulfate transporter;PDBTitle: stas domain of ychm bound to acp
15 c3lloA_
99.7
15
PDB header: motor proteinChain: A: PDB Molecule: prestin;PDBTitle: crystal structure of the stas domain of motor protein prestin (anion2 transporter slc26a5)
16 c6rtfA_
99.7
20
PDB header: membrane proteinChain: A: PDB Molecule: solute carrier family 26 member 9,solute carrier family 26PDBTitle: structure of murine solute carrier 26 family member a9 (slc26a9) anion2 transporter in an intermediate state
17 c6ic4K_
99.6
21
PDB header: protein transportChain: K: PDB Molecule: ttg2e;PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
18 c3lklB_
99.5
14
PDB header: transport proteinChain: B: PDB Molecule: antisigma-factor antagonist stas;PDBTitle: crystal structure of the c-terminal domain of anti-sigma factor2 antagonist stas from rhodobacter sphaeroides
19 c5da0A_
99.2
19
PDB header: transport proteinChain: A: PDB Molecule: sulphate transporter;PDBTitle: structure of the the slc26 transporter slc26dg in complex with a2 nanobody
20 d2q3la1
96.3
12
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Sfri0576-like
21 c3ih9A_
not modelled
95.1
20
PDB header: hydrolaseChain: A: PDB Molecule: salt-tolerant glutaminase;PDBTitle: crystal structure analysis of mglu in its tris form
22 c3bl4B_
not modelled
90.8
16
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (arth_0117) from2 arthrobacter sp. fb24 at 2.20 a resolution
23 c3bezC_
not modelled
88.9
13
PDB header: hydrolaseChain: C: PDB Molecule: protease 4;PDBTitle: crystal structure of escherichia coli signal peptide peptidase (sppa),2 semet crystals
24 c2pr7A_
not modelled
78.8
17
PDB header: hydrolaseChain: A: PDB Molecule: haloacid dehalogenase/epoxide hydrolase family;PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
25 c3rstH_
not modelled
77.0
11
PDB header: hydrolaseChain: H: PDB Molecule: signal peptide peptidase sppa;PDBTitle: crystal structure of bacillus subtilis signal peptide peptidase a
26 d2ooka1
not modelled
71.9
14
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Sfri0576-like
27 c3ghfA_
not modelled
59.0
15
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the septum site-determining protein minc from2 salmonella typhimurium
28 c3kthD_
not modelled
50.9
11
PDB header: hydrolaseChain: D: PDB Molecule: atp-dependent clp protease proteolytic subunit;PDBTitle: structure of clpp from bacillus subtilis in orthorombic crystal form
29 c2cbyG_
not modelled
48.4
15
PDB header: hydrolaseChain: G: PDB Molecule: atp-dependent clp protease proteolytic subunit 1;PDBTitle: crystal structure of the atp-dependent clp protease proteolytic2 subunit 1 (clpp1) from mycobacterium tuberculosis
30 c4yajA_
not modelled
47.7
19
PDB header: ligaseChain: A: PDB Molecule: alpha subunit of acetyl-coenzyme a synthetasePDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
31 d1tg6a1
not modelled
43.4
14
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Clp protease, ClpP subunit
32 c4umfC_
not modelled
39.6
11
PDB header: hydrolaseChain: C: PDB Molecule: 3-deoxy-d-manno-octulosonate 8-phosphate phosphatase kdsc;PDBTitle: crystal structure of 3-deoxy-d-manno-octulosonate 8-2 phosphate phosphatase from moraxella catarrhalis in3 complex with magnesium ion, phosphate ion and kdo molecule
33 c3kc2A_
not modelled
39.3
17
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ykr070w;PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
34 c2deoA_
not modelled
35.2
9
PDB header: hydrolaseChain: A: PDB Molecule: 441aa long hypothetical nfed protein;PDBTitle: 1510-n membrane protease specific for a stomatin homolog from2 pyrococcus horikoshii
35 c4eogA_
not modelled
34.7
13
PDB header: dna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of csx1 of pyrococcus furiosus
36 d2csua3
not modelled
33.9
12
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains
37 c2yx6C_
not modelled
31.6
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
38 c4olqD_
not modelled
30.9
18
PDB header: lyaseChain: D: PDB Molecule: enoyl-coa hydratase/isomerase family protein;PDBTitle: crystal structure of a putative enoyl-coa hydratase/isomerase family2 protein from hyphomonas neptunium
39 c1tg6G_
not modelled
30.6
11
PDB header: hydrolaseChain: G: PDB Molecule: putative atp-dependent clp protease proteolytic subunit;PDBTitle: crystallography and mutagenesis point to an essential role for the n-2 terminus of human mitochondrial clpp
40 d1t3va_
not modelled
28.9
15
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
41 d1o13a_
not modelled
27.6
15
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
42 d2cbya1
not modelled
27.4
19
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Clp protease, ClpP subunit
43 c2dfwA_
not modelled
25.7
19
PDB header: hydrolaseChain: A: PDB Molecule: salt-tolerant glutaminase;PDBTitle: crystal structure of a major fragment of the salt-tolerant2 glutaminase from micrococcus luteus k-3
44 d1yg6a1
not modelled
22.9
7
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Clp protease, ClpP subunit
45 c3qmjA_
not modelled
20.7
14
PDB header: lyaseChain: A: PDB Molecule: enoyl-coa hydratase, echa8_6;PDBTitle: crystal structure of enoyl-coa hydratase echa8_6 from mycobacterium2 marinum
46 c2csuB_
not modelled
20.6
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 457aa long hypothetical protein;PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
47 d1y7oa1
not modelled
19.6
11
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Clp protease, ClpP subunit
48 c2w3pB_
not modelled
19.4
11
PDB header: lyaseChain: B: PDB Molecule: benzoyl-coa-dihydrodiol lyase;PDBTitle: boxc crystal structure
49 c2mwgB_
not modelled
18.8
10
PDB header: protein bindingChain: B: PDB Molecule: blue-light photoreceptor;PDBTitle: full-length solution structure of ytva, a lov-photoreceptor protein2 and regulator of bacterial stress response
50 c2wfbA_
not modelled
18.1
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
51 c4hgnB_
not modelled
17.6
13
PDB header: hydrolaseChain: B: PDB Molecule: 2-keto-3-deoxy-d-manno-octulosonate 8-phosphatePDBTitle: crystal structure of 2-keto-3-deoxyoctulosonate 8-phosphate2 phosphohydrolase from bacteroides thetaiotaomicron
52 d1rdua_
not modelled
17.1
20
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
53 c2wmhA_
not modelled
16.9
6
PDB header: hydrolaseChain: A: PDB Molecule: fucolectin-related protein;PDBTitle: crystal structure of the catalytic module of a family 982 glycoside hydrolase from streptococcus pneumoniae tigr4 in3 complex with the h-disaccharide blood group antigen.
54 c6g4qB_
not modelled
16.6
16
PDB header: ligaseChain: B: PDB Molecule: succinate--coa ligase [adp-forming] subunit beta,PDBTitle: structure of human adp-forming succinyl-coa ligase complex suclg1-2 sucla2
55 d1eo1a_
not modelled
16.1
17
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
56 c2ej5B_
not modelled
16.1
7
PDB header: lyaseChain: B: PDB Molecule: enoyl-coa hydratase subunit ii;PDBTitle: crystal structure of gk2038 protein (enoyl-coa hydratase subunit ii)2 from geobacillus kaustophilus
57 c2hroA_
not modelled
14.8
9
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
58 d1ptma_
not modelled
13.4
10
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PdxA-like
59 c2j5iF_
not modelled
13.3
12
PDB header: lyaseChain: F: PDB Molecule: p-hydroxycinnamoyl coa hydratase/lyase;PDBTitle: crystal structure of hydroxycinnamoyl-coa hydratase-lyase
60 c6iqsD_
not modelled
13.1
5
PDB header: hydrolaseChain: D: PDB Molecule: tail-specific protease;PDBTitle: crystal structure of prc with l245a and l340g mutations in complex2 with nlpi
61 c6k4fU_
not modelled
12.6
21
PDB header: biosynthetic proteinChain: U: PDB Molecule: duf1987 domain-containing protein;PDBTitle: siac of pseudomonas aeruginosa
62 d1s1ma2
not modelled
12.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
63 c3isaA_
not modelled
11.2
29
PDB header: hydrolaseChain: A: PDB Molecule: putative enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of putative enoyl-coa hydratase/isomerase from2 bordetella parapertussis
64 c2qtdA_
not modelled
11.1
5
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein mj0327;PDBTitle: crystal structure of a putative dinitrogenase (mj0327) from2 methanocaldococcus jannaschii dsm at 1.70 a resolution
65 c4wczB_
not modelled
10.9
13
PDB header: isomeraseChain: B: PDB Molecule: enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of a putative enoyl-coa hydratase/isomerase from2 novosphingobium aromaticivorans
66 c2dgdD_
not modelled
10.8
10
PDB header: lyaseChain: D: PDB Molecule: 223aa long hypothetical arylmalonate decarboxylase;PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
67 c3re1B_
not modelled
10.8
22
PDB header: lyaseChain: B: PDB Molecule: uroporphyrinogen-iii synthetase;PDBTitle: crystal structure of uroporphyrinogen iii synthase from pseudomonas2 syringae pv. tomato dc3000
68 d2f0ca1
not modelled
10.6
15
Fold: Virus attachment protein globular domainSuperfamily: Virus attachment protein globular domainFamily: Lactophage receptor-binding protein head domain
69 d1sg4a1
not modelled
10.1
4
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Crotonase-like
70 d1tqya1
not modelled
10.0
7
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related
71 d2nu7b1
not modelled
9.9
9
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains
72 c3uosH_
not modelled
9.9
17
PDB header: ribosomeChain: H: PDB Molecule: 50s ribosomal protein l10;PDBTitle: crystal structure of release factor rf3 trapped in the gtp state on a2 rotated conformation of the ribosome (without viomycin)
73 d2bufa1
not modelled
9.7
9
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase
74 c2hwgA_
not modelled
9.6
7
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
75 d1fmfa_
not modelled
9.1
15
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
76 d1czan4
not modelled
9.0
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
77 c5udtD_
not modelled
9.0
15
PDB header: transferaseChain: D: PDB Molecule: lactate racemization operon protein lare;PDBTitle: lare, a sulfur transferase involved in synthesis of the cofactor for2 lactate racemase, in complex with amp
78 d1bg3a2
not modelled
8.9
9
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
79 c2ppyE_
not modelled
8.8
13
PDB header: lyaseChain: E: PDB Molecule: enoyl-coa hydratase;PDBTitle: crystal structure of enoyl-coa hydrates (gk_1992) from geobacillus2 kaustophilus hta426
80 d1ro5a_
not modelled
8.8
8
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Autoinducer synthetase
81 c3igxA_
not modelled
8.6
17
PDB header: transferaseChain: A: PDB Molecule: transaldolase;PDBTitle: 1.85 angstrom resolution crystal structure of transaldolase b (tala)2 from francisella tularensis.
82 c5wybB_
not modelled
8.6
18
PDB header: isomeraseChain: B: PDB Molecule: probable enoyl-coa hydratase/isomerase;PDBTitle: structure of pseudomonas aeruginosa dspi
83 c3i47A_
not modelled
8.4
11
PDB header: lyaseChain: A: PDB Molecule: enoyl coa hydratase/isomerase (crotonase);PDBTitle: crystal structure of putative enoyl coa hydratase/isomerase2 (crotonase) from legionella pneumophila subsp. pneumophila str.3 philadelphia 1
84 c2b5oA_
not modelled
8.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin--nadp reductase;PDBTitle: ferredoxin-nadp reductase
85 c3mn1B_
not modelled
8.3
14
PDB header: hydrolaseChain: B: PDB Molecule: probable yrbi family phosphatase;PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
86 c3rsiA_
not modelled
8.3
4
PDB header: isomeraseChain: A: PDB Molecule: putative enoyl-coa hydratase/isomerase;PDBTitle: the structure of a putative enoyl-coa hydratase/isomerase from2 mycobacterium abscessus atcc 19977 / dsm 44196
87 d1ys9a1
not modelled
8.3
19
Fold: HAD-likeSuperfamily: HAD-likeFamily: NagD-like
88 c6melB_
not modelled
8.2
10
PDB header: ligaseChain: B: PDB Molecule: succinate--coa ligase [adp-forming] subunit beta;PDBTitle: succinyl-coa synthase from campylobacter jejuni
89 c4es6A_
not modelled
8.1
20
PDB header: lyaseChain: A: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: crystal structure of hemd (pa5259) from pseudomonas aeruginosa (pao1)2 at 2.22 a resolution
90 c4u0gG_
not modelled
8.1
11
PDB header: hydrolase/antibioticChain: G: PDB Molecule: atp-dependent clp protease proteolytic subunit 2;PDBTitle: crystal structure of m. tuberculosis clpp1p2 bound to adep and agonist
91 c2bg5C_
not modelled
8.0
12
PDB header: transferaseChain: C: PDB Molecule: phosphoenolpyruvate-protein kinase;PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme i-domain2 from the thermoanaerobacter tengcongensis pep: sugar3 phosphotransferase system (pts)
92 c3efaA_
not modelled
7.9
18
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase;PDBTitle: crystal structure of putative n-acetyltransferase from lactobacillus2 plantarum
93 c3n1uA_
not modelled
7.7
13
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, had superfamily, subfamily iii a;PDBTitle: structure of putative had superfamily (subfamily iii a) hydrolase from2 legionella pneumophila
94 c3gkuB_
not modelled
7.6
15
PDB header: rna binding proteinChain: B: PDB Molecule: probable rna-binding protein;PDBTitle: crystal structure of a probable rna-binding protein from clostridium2 symbiosum atcc 14940
95 c2hx1D_
not modelled
7.6
8
PDB header: hydrolaseChain: D: PDB Molecule: predicted sugar phosphatases of the had superfamily;PDBTitle: crystal structure of possible sugar phosphatase, had superfamily2 (zp_00311070.1) from cytophaga hutchinsonii atcc 33406 at 2.10 a3 resolution
96 d2qale1
not modelled
7.4
22
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
97 c5ve2J_
not modelled
7.1
4
PDB header: isomerase,lyaseChain: J: PDB Molecule: enoyl-coa hydratase;PDBTitle: crystal structure of enoyl-coa hydratase/isomerase from2 pseudoalteromonas atlantica t6c at 2.3 a resolution.
98 c3trrA_
not modelled
7.0
9
PDB header: isomeraseChain: A: PDB Molecule: probable enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
99 c2q8fA_
not modelled
6.9
10
PDB header: transferaseChain: A: PDB Molecule: [pyruvate dehydrogenase [lipoamide]] kinase isozyme 1;PDBTitle: structure of pyruvate dehydrogenase kinase isoform 1