1 c5xnxB_
100.0
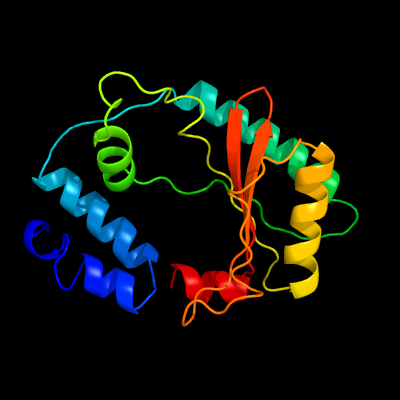
20
PDB header: hydrolase, transferaseChain: B: PDB Molecule: bifunctional (p)ppgpp synthase/hydrolase rela;PDBTitle: crystallographic structure of the enzymatically active n-terminal2 domain of the rel protein from mycobacterium tuberculosis
2 c1vj7B_
100.0
18
PDB header: hydrolase, transferaseChain: B: PDB Molecule: bifunctional rela/spot;PDBTitle: crystal structure of the bifunctional catalytic fragment of relseq,2 the rela/spot homolog from streptococcus equisimilis.
3 c5dedA_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase yjbm;PDBTitle: crystal structure of the small alarmone synthethase 1 from bacillus2 subtilis bound to its product pppgpp
4 d1vj7a2
100.0
20
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: RelA/SpoT domain
5 c6fgkA_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase ywac;PDBTitle: crystal structure of the small alarmone synthethase 2 from bacillus2 subtilis
6 c3l9dA_
100.0
21
PDB header: transferaseChain: A: PDB Molecule: putative gtp pyrophosphokinase;PDBTitle: the crystal structure of smu.1046c from streptococcus mutans ua159
7 d2be3a1
100.0
19
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: RelA/SpoT domain
8 c6ex0B_
100.0
24
PDB header: transferaseChain: B: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of relp (sas2) from staphylococcus aureus bound to2 pppgpp in the post-catalytic state
9 c3u24A_
65.7
17
PDB header: lipid binding proteinChain: A: PDB Molecule: putative lipoprotein;PDBTitle: the structure of a putative lipoprotein of unknown function from2 shewanella oneidensis.
10 d1g71a_
36.0
15
Fold: Prim-pol domainSuperfamily: Prim-pol domainFamily: PriA-like
11 c4nf9A_
29.4
12
PDB header: cell cycleChain: A: PDB Molecule: protein casc5;PDBTitle: structure of the knl1/nsl1 complex
12 c5suhB_
25.4
27
PDB header: structural proteinChain: B: PDB Molecule: msm0271 protein;PDBTitle: the structure of double ringed trimeric shell protein msm0271 from the2 rmm microcompartment
13 c4ht7G_
23.0
18
PDB header: protein bindingChain: G: PDB Molecule: co2 concentrating mechanism protein p;PDBTitle: co2 concentrating mechanism protein p, ccmp form 2
14 c4ernA_
22.2
27
PDB header: hydrolaseChain: A: PDB Molecule: tfiih basal transcription factor complex helicase xpbPDBTitle: crystal structure of the c-terminal domain of human xpb/ercc-32 excision repair protein at 1.80 a
15 c5v75E_
19.5
11
PDB header: structural proteinChain: E: PDB Molecule: microcompartments protein;PDBTitle: structure of haliangium ochraceum bmc-t ho-5816
16 c3o0yC_
18.1
24
PDB header: lipid binding proteinChain: C: PDB Molecule: lipoprotein;PDBTitle: the crystal structure of the putative lipoprotein from colwellia2 psychrerythraea
17 c5v76A_
17.1
16
PDB header: structural proteinChain: A: PDB Molecule: microcompartments protein;PDBTitle: structure of haliangium ochraceum bmc-t ho-3341
18 d2bi7a2
14.3
30
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: UDP-galactopyranose mutase
19 d1v33a_
14.0
17
Fold: Prim-pol domainSuperfamily: Prim-pol domainFamily: PriA-like
20 d1m55a_
12.4
10
Fold: Origin of replication-binding domain, RBD-likeSuperfamily: Origin of replication-binding domain, RBD-likeFamily: Replication protein Rep, nuclease domain
21 c3ohmB_
not modelled
12.2
23
PDB header: signaling protein / hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterasePDBTitle: crystal structure of activated g alpha q bound to its effector2 phospholipase c beta 3
22 c3f56F_
not modelled
12.2
16
PDB header: structural proteinChain: F: PDB Molecule: csos1d;PDBTitle: the structure of a previously undetected carboxysome shell protein:2 csos1d from prochlorococcus marinus med4
23 c1oegA_
not modelled
11.4
35
PDB header: apolipoproteinChain: A: PDB Molecule: apolipoprotein e;PDBTitle: peptide of human apoe residues 267-289, nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:sds mole ratio3 of 1:90
24 c3rmsA_
not modelled
11.2
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein svir_20580 from2 saccharomonospora viridis
25 c5l39D_
not modelled
10.9
21
PDB header: structural proteinChain: D: PDB Molecule: rmm microcompartment shell protein msm0275;PDBTitle: the structure of the fused permuted hexameric shell protein msm02752 from the rmm microcompartment
26 d1p5dx4
not modelled
10.3
23
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain
27 c6b4aB_
not modelled
9.8
16
PDB header: structural proteinChain: B: PDB Molecule: doublecortin;PDBTitle: crystal structure of the c-terminal domain of doublecortin (tgdcx)2 from toxoplasma gondii me49
28 c4abxB_
not modelled
9.7
13
PDB header: dna binding proteinChain: B: PDB Molecule: dna repair protein recn;PDBTitle: crystal structure of deinococcus radiodurans recn coiled-2 coil domain
29 c2p6nA_
not modelled
9.6
29
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase ddx41;PDBTitle: human dead-box rna helicase ddx41, helicase domain
30 d1bgva2
not modelled
9.4
17
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases
31 d1g5ta_
not modelled
8.7
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
32 c1oefA_
not modelled
8.6
33
PDB header: apolipoproteinChain: A: PDB Molecule: apolipoprotein e;PDBTitle: peptide of human apoe residues 263-286, nmr, 5 structures2 at ph 4.8, 37 degrees celsius and peptide:sds mole ratio3 of 1:90
33 c1s1i0_
not modelled
8.5
20
PDB header: ribosomeChain: 0: PDB Molecule: 60s ribosomal protein l32;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
34 c2l9mA_
not modelled
8.4
15
PDB header: apoptosis inhibitorChain: A: PDB Molecule: baculoviral iap repeat-containing protein 2;PDBTitle: structure of ciap1 card
35 d1e52a_
not modelled
7.4
21
Fold: Long alpha-hairpinSuperfamily: C-terminal UvrC-binding domain of UvrBFamily: C-terminal UvrC-binding domain of UvrB
36 c3bzjA_
not modelled
7.3
18
PDB header: hydrolaseChain: A: PDB Molecule: uv endonuclease;PDBTitle: uvde k229l
37 c6g4ww_
not modelled
7.1
22
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s15a;PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state a
38 c5h29A_
not modelled
6.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin reductase/glutathione-related protein;PDBTitle: crystal structure of the ntd_n/c domain of alkylhydroperoxide2 reductase ahpf from enterococcus faecalis (v583)
39 d1tf5a4
not modelled
6.9
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain
40 c3earA_
not modelled
6.8
23
PDB header: hydrolaseChain: A: PDB Molecule: hera;PDBTitle: novel dimerization motif in the dead box rna helicase hera: form 1,2 partial dimer
41 d1ik6a1
not modelled
6.7
15
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase Pyr module
42 d2z15a1
not modelled
6.6
29
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like
43 c1r1mA_
not modelled
6.5
50
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein class 4;PDBTitle: structure of the ompa-like domain of rmpm from neisseria2 meningitidis
44 d1r1ma_
not modelled
6.5
50
Fold: Bacillus chorismate mutase-likeSuperfamily: OmpA-likeFamily: OmpA-like
45 c4c46B_
not modelled
6.5
23
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4, general control protein gcn4;PDBTitle: andrei-n-lvpas fused to gcn4 adaptors
46 c2hjvB_
not modelled
6.3
27
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase dbpa;PDBTitle: structure of the second domain (residues 207-368) of the2 bacillus subtilis yxin protein
47 d3e9va1
not modelled
6.1
29
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like
48 c3iz5h_
not modelled
6.0
20
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l7a (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
49 d1t5ia_
not modelled
5.9
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain
50 c4uj3B_
not modelled
5.9
30
PDB header: transport proteinChain: B: PDB Molecule: rab-3a-interacting protein;PDBTitle: crystal structure of human rab11-rabin8-fip3
51 c3j21b_
not modelled
5.9
13
PDB header: ribosomeChain: B: PDB Molecule: 50s ribosomal protein l2p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
52 c2mviA_
not modelled
5.8
83
PDB header: antimicrobial proteinChain: A: PDB Molecule: bacteriocin plantarican asm1;PDBTitle: structure of the s-glycosylated bacteriocin asm1
53 c6ne3W_
not modelled
5.7
19
PDB header: dna binding protein/dnaChain: W: PDB Molecule: swi/snf-related matrix-associated actin-dependent regulatorPDBTitle: cryo-em structure of singly-bound snf2h-nucleosome complex with snf2h2 bound at shl-2
54 c3j3be_
not modelled
5.7
20
PDB header: ribosomeChain: E: PDB Molecule: 60s ribosomal protein l6;PDBTitle: structure of the human 60s ribosomal proteins
55 c4lrvL_
not modelled
5.6
40
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
56 c6ivuB_
not modelled
5.6
17
PDB header: transcriptionChain: B: PDB Molecule: rna polymerase sigma factor sigi1;PDBTitle: solution structure of the sigma-anti-sigma factor complex rsgi1n-2 sigi1c from clostridium thermocellum
57 c3zf7i_
not modelled
5.5
20
PDB header: ribosomeChain: I: PDB Molecule: 60s ribosomal protein l18;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
58 c2c1iA_
not modelled
5.5
18
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan glcnac deacetylase;PDBTitle: structure of streptococcus pneumoniae peptidoglycan deacetylase2 (sppgda) d 275 n mutant.
59 d1x93a1
not modelled
5.5
7
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
60 c1x93B_
not modelled
5.5
7
PDB header: transcriptionChain: B: PDB Molecule: hypothetical protein hp0222;PDBTitle: nmr structure of helicobacter pylori hp0222
61 d1vqoy1
not modelled
5.4
25
Fold: Barstar-likeSuperfamily: Ribosomal protein L32eFamily: Ribosomal protein L32e
62 c3cceY_
not modelled
5.4
25
PDB header: ribosomeChain: Y: PDB Molecule: 50s ribosomal protein l32e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation u2535a
63 c4ct4B_
not modelled
5.4
23
PDB header: rna binding proteinChain: B: PDB Molecule: probable atp-dependent rna helicase ddx6;PDBTitle: cnot1 mif4g domain - ddx6 complex
64 d1g64b_
not modelled
5.3
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
65 c3vcmP_
not modelled
5.1
20
PDB header: hydrolaseChain: P: PDB Molecule: prorenin;PDBTitle: crystal structure of human prorenin