| 1 |
|
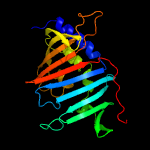
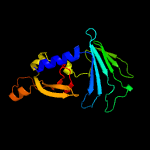
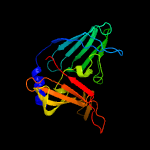
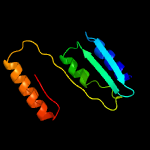
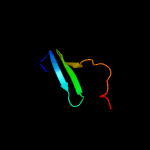
PDB 4qa8 chain A
Region: 52 - 261
Aligned: 210
Modelled: 210
Confidence: 100.0%
Identity: 100%
PDB header:lipid transport
Chain: A: PDB Molecule:putative lipoprotein lprf;
PDBTitle: crystal structure of lprf from mycobacterium bovis
Phyre2
| 2 |
|
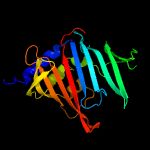
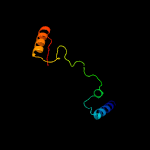
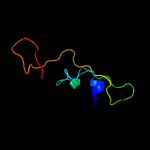
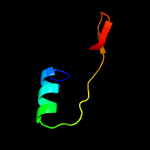
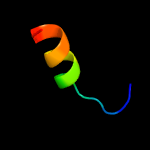
PDB 3mha chain B
Region: 53 - 254
Aligned: 191
Modelled: 202
Confidence: 100.0%
Identity: 31%
PDB header:lipid binding protein
Chain: B: PDB Molecule:lipoprotein lprg;
PDBTitle: crystal structure of lprg from mycobacterium tuberculosis bound to pim
Phyre2
| 3 |
|
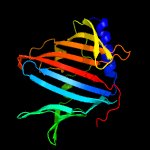
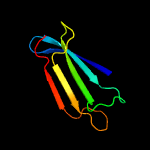
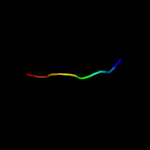
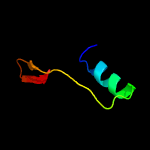
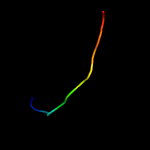
PDB 2byo chain A domain 1
Region: 54 - 257
Aligned: 181
Modelled: 198
Confidence: 100.0%
Identity: 29%
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factors
Superfamily: Prokaryotic lipoproteins and lipoprotein localization factors
Family: LppX-like
Phyre2
| 4 |
|
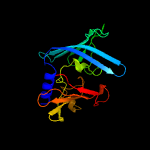
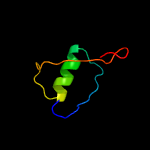
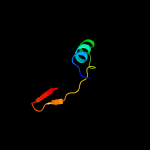
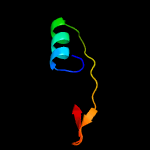
PDB 3buu chain B
Region: 57 - 250
Aligned: 171
Modelled: 194
Confidence: 97.5%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized lola superfamily protein ne2245;
PDBTitle: crystal structure of lola superfamily protein ne2245 from2 nitrosomonas europaea
Phyre2
| 5 |
|
PDB 2v43 chain A
Region: 57 - 228
Aligned: 146
Modelled: 172
Confidence: 92.3%
Identity: 14%
PDB header:regulator
Chain: A: PDB Molecule:sigma-e factor regulatory protein rseb;
PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
Phyre2
| 6 |
|
PDB 4mxt chain A
Region: 56 - 258
Aligned: 164
Modelled: 186
Confidence: 92.2%
Identity: 14%
PDB header:protein transport
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an outer-membrane lipoprotein carrier protein2 (bacuni_04723) from bacteroides uniformis atcc 8492 at 1.40 a3 resolution
Phyre2
| 7 |
|
PDB 3woa chain A
Region: 11 - 80
Aligned: 70
Modelled: 70
Confidence: 62.4%
Identity: 11%
PDB header:dna binding protein, sugar binding prote
Chain: A: PDB Molecule:repressor protein ci, maltose-binding periplasmic protein;
PDBTitle: crystal structure of lambda repressor (1-45) fused with maltose-2 binding protein
Phyre2
| 8 |
|
PDB 4z48 chain B
Region: 171 - 250
Aligned: 65
Modelled: 70
Confidence: 57.5%
Identity: 17%
PDB header:structural biology, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf1329 family protein (despig_00262) from2 desulfovibrio piger atcc 29098 at 1.75 a resolution
Phyre2
| 9 |
|
PDB 4mjs chain Q
Region: 180 - 223
Aligned: 44
Modelled: 44
Confidence: 29.4%
Identity: 16%
PDB header:transferase/protein binding
Chain: Q: PDB Molecule:protein kinase c zeta type;
PDBTitle: crystal structure of a pb1 complex
Phyre2
| 10 |
|
PDB 3bk5 chain A
Region: 58 - 250
Aligned: 177
Modelled: 193
Confidence: 8.9%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative outer membrane lipoprotein-sorting protein;
PDBTitle: crystal structure of putative outer membrane lipoprotein-sorting2 protein domain from vibrio parahaemolyticus
Phyre2
| 11 |
|
PDB 3a5z chain D
Region: 191 - 257
Aligned: 54
Modelled: 58
Confidence: 8.8%
Identity: 20%
PDB header:ligase
Chain: D: PDB Molecule:elongation factor p;
PDBTitle: crystal structure of escherichia coli genx in complex with elongation2 factor p
Phyre2
| 12 |
|
PDB 1wmx chain A
Region: 212 - 219
Aligned: 8
Modelled: 8
Confidence: 8.8%
Identity: 50%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 30 carbohydrate binding module, CBM30 (PKD repeat)
Phyre2
| 13 |
|
PDB 3ks6 chain A
Region: 193 - 224
Aligned: 32
Modelled: 32
Confidence: 7.8%
Identity: 6%
PDB header:hydrolase
Chain: A: PDB Molecule:glycerophosphoryl diester phosphodiesterase;
PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (17743486) from agrobacterium tumefaciens str. c583 (dupont) at 1.80 a resolution
Phyre2
| 14 |
|
PDB 5cw9 chain A
Region: 159 - 248
Aligned: 90
Modelled: 90
Confidence: 6.6%
Identity: 13%
PDB header:de novo protein
Chain: A: PDB Molecule:de novo designed ferredoxin-ferredoxin domain insertion
PDBTitle: crystal structure of de novo designed ferredoxin-ferredoxin domain2 insertion protein
Phyre2
| 15 |
|
PDB 2o55 chain A
Region: 193 - 224
Aligned: 32
Modelled: 32
Confidence: 6.4%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative glycerophosphodiester phosphodiesterase;
PDBTitle: crystal structure of a putative glycerophosphodiester2 phosphodiesterase from galdieria sulphuraria
Phyre2
| 16 |
|
PDB 4oec chain D
Region: 193 - 224
Aligned: 32
Modelled: 32
Confidence: 5.6%
Identity: 6%
PDB header:hydrolase
Chain: D: PDB Molecule:glycerophosphoryl diester phosphodiesterase;
PDBTitle: crystal structure of glycerophosphodiester phosphodiesterase from2 thermococcus kodakarensis kod1
Phyre2
| 17 |
|
PDB 3qvq chain B
Region: 193 - 224
Aligned: 32
Modelled: 32
Confidence: 5.4%
Identity: 6%
PDB header:hydrolase
Chain: B: PDB Molecule:phosphodiesterase olei02445;
PDBTitle: the structure of an oleispira antarctica phosphodiesterase olei024452 in complex with the product sn-glycerol-3-phosphate
Phyre2
| 18 |
|
PDB 3n3f chain B
Region: 117 - 153
Aligned: 37
Modelled: 37
Confidence: 5.4%
Identity: 11%
PDB header:protein binding
Chain: B: PDB Molecule:collagen alpha-1(xv) chain;
PDBTitle: crystal structure of the human collagen xv trimerization domain: a2 potent trimerizing unit common to multiplexin collagens
Phyre2
| 19 |
|
PDB 2hcu chain A
Region: 9 - 23
Aligned: 15
Modelled: 15
Confidence: 5.2%
Identity: 27%
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus mutans
Phyre2
| 20 |
|
PDB 1k81 chain A
Region: 7 - 18
Aligned: 12
Modelled: 12
Confidence: 5.2%
Identity: 50%
Fold: Zinc-binding domain of translation initiation factor 2 beta
Superfamily: Zinc-binding domain of translation initiation factor 2 beta
Family: Zinc-binding domain of translation initiation factor 2 beta
Phyre2