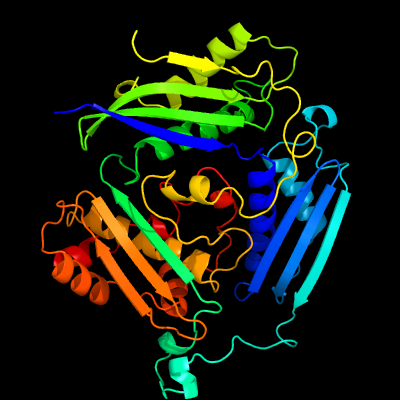
| 1 | c5h9uC_
|
|
 |
100.0 |
59 |
PDB header:transferase
Chain: C: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of a thermostable methionine adenosyltransferase
|
|
|
|
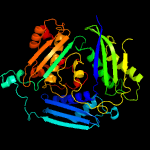
| 2 | c1rg9D_
|
|
 |
100.0 |
61 |
PDB header:transferase
Chain: D: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: s-adenosylmethionine synthetase complexed with sam and ppnp
|
|
|
|
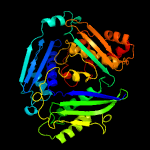
| 3 | c3rv2B_
|
|
 |
100.0 |
95 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
|
|
|
|
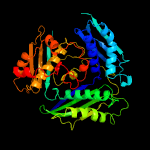
| 4 | c2obvA_
|
|
 |
100.0 |
55 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthetase isoform type-1;
PDBTitle: crystal structure of the human s-adenosylmethionine synthetase 1 in2 complex with the product
|
|
|
|
| 5 | c4odjA_
|
|
 |
100.0 |
51 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of a putative s-adenosylmethionine synthetase from2 cryptosporidium hominis in complex with s-adenosyl-methionine
|
|
|
|
| 6 | c3so4C_
|
|
 |
100.0 |
52 |
PDB header:transferase
Chain: C: PDB Molecule:methionine-adenosyltransferase;
PDBTitle: methionine-adenosyltransferase from entamoeba histolytica
|
|
|
|
| 7 | c3imlB_
|
|
 |
100.0 |
60 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from burkholderia2 pseudomallei
|
|
|
|
| 8 | c4le5A_
|
|
 |
100.0 |
43 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: structure of an unusual s-adenosylmethionine synthetase from2 campylobacter jejuni
|
|
|
|
| 9 | d1mxaa3
|
|
 |
100.0 |
67 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 10 | d1qm4a3
|
|
 |
100.0 |
62 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 11 | d2p02a3
|
|
 |
100.0 |
61 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 12 | d1mxaa2
|
|
 |
100.0 |
50 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 13 | d2p02a2
|
|
 |
100.0 |
45 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 14 | d1qm4a2
|
|
 |
100.0 |
49 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 15 | d2p02a1
|
|
 |
100.0 |
56 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 16 | d1mxaa1
|
|
 |
100.0 |
60 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 17 | d1qm4a1
|
|
 |
100.0 |
56 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
|
|
|
| 18 | c4hpvA_
|
|
 |
99.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from sulfolobus2 solfataricus
|
|
|
|
| 19 | c4l4qA_
|
|
 |
99.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: methionine adenosyltransferase
|
|
|
|
| 20 | c2mblA_
|
|
 |
52.2 |
21 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7 fold protein top7m13;
PDBTitle: solution nmr structure of de novo designed top7 fold protein top7m13,2 northeast structural genomics consortium (nesg) target or33
|
|
|
|
| 21 | d1w9ha1 |
|
not modelled |
52.0 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
|
|
| 22 | c4idxA_ |
|
not modelled |
44.5 |
43 |
PDB header:dna binding protein
Chain: A: PDB Molecule:nucleocapsid protein;
PDBTitle: hexameric crystal structure of schmallenberg virus nucleoprotein
|
|
|
| 23 | d1u04a2 |
|
not modelled |
39.8 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
|
|
| 24 | c3ho1A_ |
|
not modelled |
38.6 |
25 |
PDB header:nucleic acid binding protein/dna/rna
Chain: A: PDB Molecule:argonaute;
PDBTitle: crystal structure of t. thermophilus argonaute n546 mutant protein2 complexed with dna guide strand and 12-nt rna target strand
|
|
|
| 25 | c5guhA_ |
|
not modelled |
37.6 |
18 |
PDB header:hydrolase/rna
Chain: A: PDB Molecule:piwi;
PDBTitle: crystal structure of silkworm piwi-clade argonaute siwi bound to pirna
|
|
|
| 26 | c4ijsB_ |
|
not modelled |
37.4 |
38 |
PDB header:rna binding protein/rna
Chain: B: PDB Molecule:nucleoprotein;
PDBTitle: crystal structure of nucleocapsid protein encoded by the prototypic2 member of orthobunyavirus
|
|
|
| 27 | d1twfc1 |
|
not modelled |
36.6 |
21 |
Fold:DCoH-like
Superfamily:RBP11-like subunits of RNA polymerase
Family:RNA polymerase alpha subunit dimerisation domain |
|
|
| 28 | c4bhhZ_ |
|
not modelled |
36.6 |
48 |
PDB header:viral protein/rna
Chain: Z: PDB Molecule:nucleoprotein;
PDBTitle: crystal structure of tetramer of la crosse virus nucleoprotein in2 complex with ssrna
|
|
|
| 29 | d1kjna_ |
|
not modelled |
34.3 |
29 |
Fold:Hypothetical protein MTH777 (MT0777)
Superfamily:Hypothetical protein MTH777 (MT0777)
Family:Hypothetical protein MTH777 (MT0777) |
|
|
| 30 | c2z51A_ |
|
not modelled |
34.0 |
35 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 2, chloroplast;
PDBTitle: crystal structure of arabidopsis cnfu involved in iron-sulfur cluster2 biosynthesis
|
|
|
| 31 | c2w42A_ |
|
not modelled |
33.7 |
16 |
PDB header:protein/dna complex
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the structure of a piwi protein from archaeoglobus fulgidus2 complexed with a 16nt dna duplex.
|
|
|
| 32 | c2w8iG_ |
|
not modelled |
31.4 |
28 |
PDB header:membrane protein
Chain: G: PDB Molecule:putative outer membrane lipoprotein wza;
PDBTitle: crystal structure of wza24-345.
|
|
|
| 33 | c3lvtA_ |
|
not modelled |
31.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hydrolase, family 38;
PDBTitle: the crystal structure of a protein in the glycosyl hydrolase family 382 from enterococcus faecalis to 2.55a
|
|
|
| 34 | c4j1jA_ |
|
not modelled |
29.5 |
33 |
PDB header:viral protein/dna
Chain: A: PDB Molecule:nucleocapsid;
PDBTitle: leanyer orthobunyavirus nucleoprotein-ssdna complex
|
|
|
| 35 | c2mraA_ |
|
not modelled |
29.2 |
23 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo designed protein or459;
PDBTitle: solution nmr structure of de novo designed protein, northeast2 structural genomics consortium (nesg) target or459
|
|
|
| 36 | c2gjhA_ |
|
not modelled |
24.5 |
21 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein;
PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
|
|
|
| 37 | d1to0a_ |
|
not modelled |
24.5 |
50 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
|
|
| 38 | c3k8zD_ |
|
not modelled |
23.9 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nad-specific glutamate dehydrogenase;
PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
|
|
|
| 39 | d1vh0a_ |
|
not modelled |
23.6 |
63 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
|
|
| 40 | c2inrA_ |
|
not modelled |
23.2 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase 4 subunit a;
PDBTitle: crystal structure of a 59 kda fragment of topoisomerase iv subunit a2 (grla) from staphylococcus aureus
|
|
|
| 41 | d2fm8a1 |
|
not modelled |
23.2 |
28 |
Fold:Secretion chaperone-like
Superfamily:Type III secretory system chaperone-like
Family:Type III secretory system chaperone |
|
|
| 42 | d1ns5a_ |
|
not modelled |
22.6 |
40 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
|
|
| 43 | c3kxyV_ |
|
not modelled |
22.3 |
54 |
PDB header:chaperone/transcription inhibitor
Chain: V: PDB Molecule:exse;
PDBTitle: crystal structure of the exsc-exse complex
|
|
|
| 44 | d1o6da_ |
|
not modelled |
22.1 |
50 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
|
|
| 45 | d2nrqa1 |
|
not modelled |
21.9 |
28 |
Fold:RL5-like
Superfamily:RL5-like
Family:SSO1042-like |
|
|
| 46 | d1vkha_ |
|
not modelled |
21.3 |
19 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Putative serine hydrolase Ydr428c |
|
|
| 47 | c2vofA_ |
|
not modelled |
20.6 |
4 |
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-related protein a1;
PDBTitle: structure of mouse a1 bound to the puma bh3-domain
|
|
|
| 48 | c5lqwL_ |
|
not modelled |
20.4 |
43 |
PDB header:splicing
Chain: L: PDB Molecule:pre-mrna-splicing factor cwc26;
PDBTitle: yeast activated spliceosome
|
|
|
| 49 | c2jvfA_ |
|
not modelled |
19.9 |
9 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
|
|
|
| 50 | d1mp9a1 |
|
not modelled |
19.7 |
20 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 51 | c4v19T_ |
|
not modelled |
19.6 |
22 |
PDB header:ribosome
Chain: T: PDB Molecule:mitoribosomal protein bl19m, mrpl19;
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
|
|
|
| 52 | c5i4aC_ |
|
not modelled |
18.8 |
18 |
PDB header:rna binding protein/rna
Chain: C: PDB Molecule:argonaute protein;
PDBTitle: x-ray crystal structure of marinitoga piezophila argonaute in complex2 with 5' oh guide rna
|
|
|
| 53 | c2mcdA_ |
|
not modelled |
18.3 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:murine norovirus 1;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus ns1/2 d94e mutant
|
|
|
| 54 | c4e0iA_ |
|
not modelled |
18.0 |
69 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mitochondrial fad-linked sulfhydryl oxidase erv1;
PDBTitle: crystal structure of the c30s/c133s mutant of erv1 from saccharomyces2 cerevisiae
|
|
|
| 55 | c5zyoD_ |
|
not modelled |
18.0 |
50 |
PDB header:transferase
Chain: D: PDB Molecule:ribosomal rna large subunit methyltransferase h;
PDBTitle: crystal structure of domain-swapped circular-permuted ybea (cp74) from2 escherichia coli
|
|
|
| 56 | d1azpa_ |
|
not modelled |
17.9 |
40 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:"Histone-like" proteins from archaea |
|
|
| 57 | d2fiqa1 |
|
not modelled |
17.8 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
|
|
| 58 | c5vogA_ |
|
not modelled |
17.4 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphoribosyltransferase;
PDBTitle: crystal structure of a hypothetical protein from neisseria gonorrhoeae2 with bound ppgpp
|
|
|
| 59 | d1aisa2 |
|
not modelled |
17.4 |
13 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 60 | c1u04A_ |
|
not modelled |
17.4 |
31 |
PDB header:hydrolase/gene regulation
Chain: A: PDB Molecule:hypothetical protein pf0537;
PDBTitle: crystal structure of full length argonaute from pyrococcus furiosus
|
|
|
| 61 | c5awhA_ |
|
not modelled |
17.2 |
21 |
PDB header:rna binding protein/dna/rna
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: rhodobacter sphaeroides argonaute in complex with guide rna/target dna2 heteroduplex
|
|
|
| 62 | c3d4oA_ |
|
not modelled |
16.6 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
|
|
| 63 | c2z8uQ_ |
|
not modelled |
16.4 |
27 |
PDB header:transcription
Chain: Q: PDB Molecule:tata-box-binding protein;
PDBTitle: methanococcus jannaschii tbp
|
|
|
| 64 | c2jkzB_ |
|
not modelled |
15.3 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: saccharomyces cerevisiae hypoxanthine-guanine2 phosphoribosyltransferase in complex with gmp (guanosine 5'-3 monophosphate) (orthorhombic crystal form)
|
|
|
| 65 | d1q33a_ |
|
not modelled |
15.2 |
60 |
Fold:Nudix
Superfamily:Nudix
Family:MutT-like |
|
|
| 66 | c5theA_ |
|
not modelled |
15.2 |
24 |
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the c-terminal lobe of a budding yeast argonaute
|
|
|
| 67 | c5vidG_ |
|
not modelled |
15.0 |
18 |
PDB header:toxin
Chain: G: PDB Molecule:bot.0671.2;
PDBTitle: receptor binding domain of bont/b in complex with mini-protein binder2 bot.0671.2
|
|
|
| 68 | c5vidI_ |
|
not modelled |
15.0 |
18 |
PDB header:toxin
Chain: I: PDB Molecule:bot.0671.2;
PDBTitle: receptor binding domain of bont/b in complex with mini-protein binder2 bot.0671.2
|
|
|
| 69 | c3aoeC_ |
|
not modelled |
15.0 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of hetero-hexameric glutamate dehydrogenase from2 thermus thermophilus (leu bound form)
|
|
|
| 70 | c5g5tA_ |
|
not modelled |
14.8 |
33 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:argonaute;
PDBTitle: structure of the argonaute protein from methanocaldcoccus janaschii in2 complex with guide dna
|
|
|
| 71 | c2mckA_ |
|
not modelled |
14.5 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:polyprotein;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
|
|
|
| 72 | d2bgxa2 |
|
not modelled |
14.5 |
21 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
|
|
| 73 | c5ijzH_ |
|
not modelled |
14.4 |
24 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nadp-specific glutamate dehydrogenase;
PDBTitle: crystal strcuture of glutamate dehydrogenase(gdh) from corynebacterium2 glutamicum
|
|
|
| 74 | c3aogA_ |
|
not modelled |
14.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase;
PDBTitle: crystal structure of glutamate dehydrogenase (gdhb) from thermus2 thermophilus (glu bound form)
|
|
|
| 75 | d1csna_ |
|
not modelled |
14.0 |
15 |
Fold:Protein kinase-like (PK-like)
Superfamily:Protein kinase-like (PK-like)
Family:Protein kinases, catalytic subunit |
|
|
| 76 | c6qzkA_ |
|
not modelled |
14.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:clostridium butyricum argonaute;
PDBTitle: structure of clostridium butyricum argonaute bound to a guide dna (5'2 deoxycytidine) and a 19-mer target dna
|
|
|
| 77 | d2f9wa2 |
|
not modelled |
13.6 |
43 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:CoaX-like |
|
|
| 78 | d1t0ga_ |
|
not modelled |
13.4 |
39 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Steroid-binding domain |
|
|
| 79 | c4m6iA_ |
|
not modelled |
13.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan amidase rv3717;
PDBTitle: structure of the reduced, zn-bound form of mycobacterium tuberculosis2 peptidoglycan amidase rv3717
|
|
|
| 80 | c1qysA_ |
|
not modelled |
13.2 |
29 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
|
|
| 81 | c2j58G_ |
|
not modelled |
13.2 |
27 |
PDB header:membrane protein
Chain: G: PDB Molecule:outer membrane lipoprotein wza;
PDBTitle: the structure of wza
|
|
|
| 82 | d1knga_ |
|
not modelled |
13.2 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
|
|
| 83 | c1v57A_ |
|
not modelled |
12.9 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbg;
PDBTitle: crystal structure of the disulfide bond isomerase dsbg
|
|
|
| 84 | d1nh2a2 |
|
not modelled |
12.8 |
20 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 85 | c5b3kA_ |
|
not modelled |
12.7 |
19 |
PDB header:electron transport
Chain: A: PDB Molecule:uncharacterized protein pa3435;
PDBTitle: c101a mutant of flavodoxin from pseudomonas aeruginosa
|
|
|
| 86 | c2bmaA_ |
|
not modelled |
12.5 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase (nadp+);
PDBTitle: the crystal structure of plasmodium falciparum glutamate2 dehydrogenase, a putative target for novel antimalarial3 drugs
|
|
|
| 87 | c5b54D_ |
|
not modelled |
12.2 |
28 |
PDB header:transferase
Chain: D: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of hydrogen sulfide-producing enzyme (fn1055) from2 fusobacterium nucleatum: lysine-dimethylated form
|
|
|
| 88 | d1yvua2 |
|
not modelled |
12.1 |
6 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:PIWI domain |
|
|
| 89 | c1wnkA_ |
|
not modelled |
12.0 |
43 |
PDB header:transcription
Chain: A: PDB Molecule:fibroin-modulator-binding-protein-1;
PDBTitle: nmr structure of fmbp-1 tandem repeat 3 in 30%(v/v) tfe2 solution
|
|
|
| 90 | c5ylyB_ |
|
not modelled |
11.9 |
6 |
PDB header:flavoprotein
Chain: B: PDB Molecule:nitrate reductase;
PDBTitle: crystal structure of the cytochrome b5 reductase domain of ulva2 prolifera nitrate reductase
|
|
|
| 91 | c2wl2B_ |
|
not modelled |
11.8 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit a;
PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
|
|
|
| 92 | c6qt9Y_ |
|
not modelled |
11.8 |
37 |
PDB header:virus
Chain: Y: PDB Molecule:orf 31;
PDBTitle: cryo-em structure of sh1 full particle.
|
|
|
| 93 | d1aisa1 |
|
not modelled |
11.8 |
29 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
|
|
| 94 | c4ediC_ |
|
not modelled |
11.7 |
17 |
PDB header:transport protein
Chain: C: PDB Molecule:ethanolamine utilization protein;
PDBTitle: disulfide bonded eutl from clostridium perfringens
|
|
|
| 95 | c2hnbA_ |
|
not modelled |
11.6 |
15 |
PDB header:electron transport
Chain: A: PDB Molecule:protein mioc;
PDBTitle: solution structure of a bacterial holo-flavodoxin
|
|
|
| 96 | d1mtyd_ |
|
not modelled |
11.3 |
59 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ribonucleotide reductase-like |
|
|
| 97 | c3u27D_ |
|
not modelled |
11.3 |
34 |
PDB header:structural protein
Chain: D: PDB Molecule:microcompartments protein;
PDBTitle: crystal structure of ethanolamine utilization protein eutl from2 leptotrichia buccalis c-1013-b
|
|
|
| 98 | c6cp8D_ |
|
not modelled |
11.3 |
39 |
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:cdii;
PDBTitle: contact-dependent growth inhibition toxin-immunity protein complex2 from from e. coli 3006
|
|
|
| 99 | c4fmoB_ |
|
not modelled |
11.1 |
32 |
PDB header:hydrolase
Chain: B: PDB Molecule:dna mismatch repair protein pms1;
PDBTitle: structure of the c-terminal domain of the saccharomyces cerevisiae2 mutl alpha (mlh1/pms1) heterodimer bound to a fragment of exo1
|
|
|