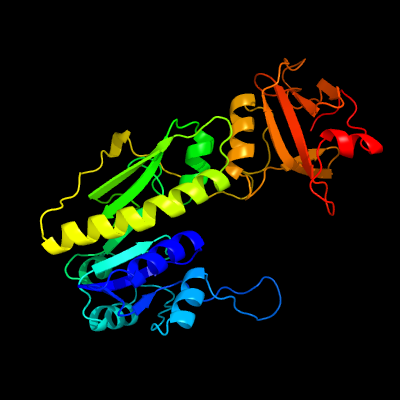
| 1 | c5uaiA_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methionyl-trna formyltransferase from pseudomonas2 aeruginosa
|
|
|
|
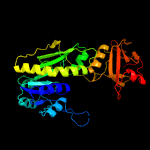
| 2 | c3rfoA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
|
|
|
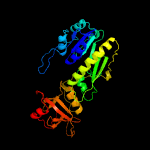
| 3 | c1fmtA_
|
|
 |
100.0 |
36 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
|
|
|
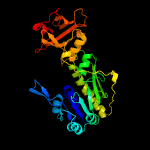
| 4 | c3q0iA_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from vibrio cholerae
|
|
|
|
| 5 | c3tqqA_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: structure of the methionyl-trna formyltransferase (fmt) from coxiella2 burnetii
|
|
|
|
| 6 | c1z7eC_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
|
|
|
| 7 | c1yrwA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
|
|
|
| 8 | c1s3iA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: crystal structure of the n terminal hydrolase domain of 10-2 formyltetrahydrofolate dehydrogenase
|
|
|
|
| 9 | c4lxuB_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:wlard, a sugar 3n-formyl transferase;
PDBTitle: dtdp-fuc3n and 5-n-formyl-thf
|
|
|
|
| 10 | c5vytD_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: D: PDB Molecule:gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-
PDBTitle: crystal structure of the wbkc n-formyltransferase (f142a variant) from2 brucella melitensis
|
|
|
|
| 11 | c5uimB_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:formyltransferase;
PDBTitle: x-ray structure of the fdtf n-formyltransferase from salmonella2 enteric o60 in complex with folinic acid and tdp-qui3n
|
|
|
|
| 12 | d2blna2
|
|
 |
100.0 |
30 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 13 | d1fmta2
|
|
 |
100.0 |
40 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 14 | d1s3ia2
|
|
 |
100.0 |
26 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 15 | d2bw0a2
|
|
 |
100.0 |
26 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 16 | c4nv1D_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:formyltransferase;
PDBTitle: crystal structure of a 4-n formyltransferase from francisella2 tularensis
|
|
|
|
| 17 | c4yfvA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:viof;
PDBTitle: x-ray structure of the 4-n-formyltransferase viof from providencia2 alcalifaciens o30
|
|
|
|
| 18 | c1zghA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from clostridium thermocellum
|
|
|
|
| 19 | c4pzuF_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: F: PDB Molecule:uncharacterized protein rv3404c/mt3512;
PDBTitle: crystal structure of a putative uncharacterize protein rv3404c and2 likely sugar n-formyltransferase from mycobacterium tuberculosis
|
|
|
|
| 20 | c6nbpA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:n-formyltransferase;
PDBTitle: crystal structure of a sugar n-formyltransferase from the plant2 pathogen pantoea ananatis
|
|
|
|
| 21 | c3kcqA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
|
|
| 22 | d1jkxa_ |
|
not modelled |
100.0 |
20 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 23 | c2ywrA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex aeolicus
|
|
|
| 24 | c3dcjA_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide formyltransferase
PDBTitle: crystal structure of glycinamide formyltransferase (purn) from2 mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-3 tetrahydrofolic acid derivative
|
|
|
| 25 | c3aufA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:glycinamide ribonucleotide transformylase 1;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 symbiobacterium toebii
|
|
|
| 26 | c3tqrA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
|
|
| 27 | c3p9xB_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
|
|
| 28 | c3av3A_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 geobacillus kaustophilus
|
|
|
| 29 | c4s1nA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
|
|
|
| 30 | d1meoa_ |
|
not modelled |
100.0 |
22 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 31 | c4ds3A_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 brucella melitensis
|
|
|
| 32 | c6ci2A_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:formyltransferase psej;
PDBTitle: crystal structure of the formyltransferase psej from anoxybacillus2 kamchatkensis
|
|
|
| 33 | c5cjjA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
| 34 | c3n0vD_ |
|
not modelled |
100.0 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
|
|
| 35 | c3w7bB_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase from thermus2 thermophilus hb8
|
|
|
| 36 | c3o1lB_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
|
|
| 37 | c3louB_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
|
|
| 38 | c3obiC_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
|
|
| 39 | c3nrbD_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
|
|
| 40 | c4xd0A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:tdp-3-aminoquinovose-n-formyltransferase;
PDBTitle: x-ray structure of the n-formyltransferase qdtf from providencia2 alcalifaciens
|
|
|
| 41 | d1zgha2 |
|
not modelled |
100.0 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 42 | d1fmta1 |
|
not modelled |
99.9 |
30 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
|
|
| 43 | d2blna1 |
|
not modelled |
99.9 |
22 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
|
|
| 44 | d2bw0a1 |
|
not modelled |
99.9 |
20 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
|
|
| 45 | d1s3ia1 |
|
not modelled |
99.9 |
22 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
|
|
| 46 | c6culG_ |
|
not modelled |
96.5 |
27 |
PDB header:transferase
Chain: G: PDB Molecule:pyoverdine synthetase f;
PDBTitle: pvdf of pyoverdin biosynthesis is a structurally unique n10-2 formyltetrahydrofolate-dependent formyltransferase
|
|
|
| 47 | c5b3uB_ |
|
not modelled |
96.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 48 | c3ezyB_ |
|
not modelled |
96.4 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from thermotoga2 maritima
|
|
|
| 49 | c3euwB_ |
|
not modelled |
96.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
|
|
| 50 | c3db2C_ |
|
not modelled |
96.3 |
12 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
|
|
| 51 | c6norB_ |
|
not modelled |
96.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nad dependent dehydrogenase;
PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
|
|
|
| 52 | c3rbvA_ |
|
not modelled |
96.0 |
20 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
|
|
| 53 | c5uibA_ |
|
not modelled |
95.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase protein;
PDBTitle: crystal structure of an oxidoreductase from agrobacterium radiobacter2 in complex with nad+, l-tartaric acid and magnesium
|
|
|
| 54 | c4hktA_ |
|
not modelled |
95.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol 2-dehydrogenase;
PDBTitle: crystal structure of a putative myo-inositol dehydrogenase from2 sinorhizobium meliloti 1021 (target psi-012312)
|
|
|
| 55 | c4mkzA_ |
|
not modelled |
95.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol dehydrogenase;
PDBTitle: crystal structure of apo scyllo-inositol dehydrogenase from2 lactobacillus casei at 77k
|
|
|
| 56 | c2ho3D_ |
|
not modelled |
95.6 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
|
|
| 57 | c3ec7C_ |
|
not modelled |
95.6 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
|
|
| 58 | c4hadD_ |
|
not modelled |
95.5 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
|
|
|
| 59 | d1zgha1 |
|
not modelled |
95.5 |
15 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
|
|
| 60 | c3e9mC_ |
|
not modelled |
95.4 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 61 | c2ynmD_ |
|
not modelled |
95.3 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
|
|
|
| 62 | c2glxD_ |
|
not modelled |
95.2 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
|
|
| 63 | c3e18A_ |
|
not modelled |
95.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
|
|
| 64 | c3nt5B_ |
|
not modelled |
95.1 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
|
|
| 65 | c4miyB_ |
|
not modelled |
95.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from lactobacillus2 casei in complex with nad and myo-inositol
|
|
|
| 66 | c3eywA_ |
|
not modelled |
94.9 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
|
|
| 67 | d1lssa_ |
|
not modelled |
94.9 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
|
|
| 68 | c3moiA_ |
|
not modelled |
94.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
|
|
| 69 | c3wgzB_ |
|
not modelled |
94.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:meso-diaminopimelate dehydrogenase;
PDBTitle: crystal structure of meso-dapdh q154l/t173i/r199m/p248s/h249n/n276s2 mutant with d-leucine of from clostridium tetani e88
|
|
|
| 70 | c3ceaA_ |
|
not modelled |
94.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
|
|
| 71 | c2ynmC_ |
|
not modelled |
94.6 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
|
|
|
| 72 | c3q2kB_ |
|
not modelled |
94.5 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
|
|
| 73 | c2q4eB_ |
|
not modelled |
94.4 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
|
|
| 74 | c2o48X_ |
|
not modelled |
94.4 |
23 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
|
|
| 75 | c3v5nA_ |
|
not modelled |
94.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
|
|
| 76 | d1qh8b_ |
|
not modelled |
94.3 |
16 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 77 | c3nklA_ |
|
not modelled |
94.2 |
14 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
|
|
| 78 | c4amuB_ |
|
not modelled |
94.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine carbamoyltransferase, catabolic;
PDBTitle: structure of ornithine carbamoyltransferase from mycoplasma2 penetrans with a p321 space group
|
|
|
| 79 | d1pvva2 |
|
not modelled |
94.0 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 80 | c1zh8B_ |
|
not modelled |
93.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
|
|
| 81 | d1a9xa4 |
|
not modelled |
93.8 |
13 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 82 | d1kewa_ |
|
not modelled |
93.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 83 | c3m2tA_ |
|
not modelled |
93.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
|
|
| 84 | d1m1nb_ |
|
not modelled |
93.7 |
18 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 85 | d1miob_ |
|
not modelled |
93.7 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 86 | c3uuwB_ |
|
not modelled |
93.7 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase with nad(p)-binding rossmann-fold
PDBTitle: 1.63 angstrom resolution crystal structure of dehydrogenase (mvim)2 from clostridium difficile.
|
|
|
| 87 | c3wb9A_ |
|
not modelled |
93.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diaminopimelate dehydrogenase;
PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
|
|
|
| 88 | c3ic5A_ |
|
not modelled |
93.6 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
|
|
| 89 | c4g65A_ |
|
not modelled |
93.6 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:trk system potassium uptake protein trka;
PDBTitle: potassium transporter peripheral membrane component (trka) from vibrio2 vulnificus
|
|
|
| 90 | c5z2fA_ |
|
not modelled |
93.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
|
|
|
| 91 | d1ydwa1 |
|
not modelled |
93.3 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 92 | d1vlva2 |
|
not modelled |
93.2 |
20 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 93 | c5kojD_ |
|
not modelled |
93.2 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase femo beta subunit protein nifk;
PDBTitle: nitrogenase mofep protein in the ids oxidized state
|
|
|
| 94 | c3l4bG_ |
|
not modelled |
93.2 |
12 |
PDB header:transport protein
Chain: G: PDB Molecule:trka k+ channel protien tm1088b;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
|
|
| 95 | c4gmfD_ |
|
not modelled |
93.2 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:yersiniabactin biosynthetic protein ybtu;
PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
|
|
|
| 96 | c2axqA_ |
|
not modelled |
93.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
|
|
| 97 | c1ofgF_ |
|
not modelled |
93.1 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
|
|
| 98 | c3fd8A_ |
|
not modelled |
93.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 99 | c1h6dL_ |
|
not modelled |
93.0 |
15 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
|
|
| 100 | c1drwA_ |
|
not modelled |
93.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
|
|
| 101 | c4gqaC_ |
|
not modelled |
92.9 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad binding oxidoreductase;
PDBTitle: crystal structure of nad binding oxidoreductase from klebsiella2 pneumoniae
|
|
|
| 102 | c5yabD_ |
|
not modelled |
92.9 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:scyllo-inositol dehydrogenase with l-glucose dehydrogenase
PDBTitle: crystal structure of scyllo-inositol dehydrogenase with l-glucose2 dehydrogenase activity
|
|
|
| 103 | c3ijpA_ |
|
not modelled |
92.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
|
|
|
| 104 | d1ml4a2 |
|
not modelled |
92.8 |
13 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 105 | c5n6yD_ |
|
not modelled |
92.7 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase vanadium-iron protein alpha chain;
PDBTitle: azotobacter vinelandii vanadium nitrogenase
|
|
|
| 106 | c6g1mA_ |
|
not modelled |
92.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: amine dehydrogenase from petrotoga mobilis; open and closed form
|
|
|
| 107 | c2p5uC_ |
|
not modelled |
92.7 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-epimerase2 complex with nad
|
|
|
| 108 | d1qh8a_ |
|
not modelled |
92.6 |
12 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 109 | d1m1na_ |
|
not modelled |
92.4 |
12 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 110 | c2p2sA_ |
|
not modelled |
92.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
|
|
|
| 111 | c3evnA_ |
|
not modelled |
92.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
|
|
|
| 112 | c5wolA_ |
|
not modelled |
91.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase dapb from coxiella2 burnetii
|
|
|
| 113 | c2pk3B_ |
|
not modelled |
91.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
|
|
| 114 | c3pdiG_ |
|
not modelled |
91.8 |
12 |
PDB header:protein binding
Chain: G: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nife;
PDBTitle: precursor bound nifen
|
|
|
| 115 | c5ugjC_ |
|
not modelled |
91.8 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
|
|
|
| 116 | c5kt0A_ |
|
not modelled |
91.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 117 | c2ph5A_ |
|
not modelled |
91.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
|
|
| 118 | c3aerC_ |
|
not modelled |
91.4 |
9 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
|
|
|
| 119 | c3kuxA_ |
|
not modelled |
91.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
|
|
|
| 120 | c5a06E_ |
|
not modelled |
91.1 |
14 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:aldose-aldose oxidoreductase;
PDBTitle: crystal structure of aldose-aldose oxidoreductase from2 caulobacter crescentus complexed with sorbitol
|
|
|