| 1 | c4e0fB_
|
|
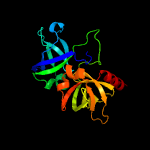 |
100.0 |
37 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase subunit alpha;
PDBTitle: crystallographic structure of trimeric riboflavin synthase from2 brucella abortus in complex with riboflavin
|
|
|
|
| 2 | c1kzlA_
|
|
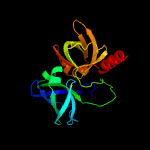 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase;
PDBTitle: riboflavin synthase from s.pombe bound to2 carboxyethyllumazine
|
|
|
|
| 3 | c1i8dB_
|
|
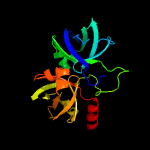 |
100.0 |
36 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase;
PDBTitle: crystal structure of riboflavin synthase
|
|
|
|
| 4 | c3a35B_
|
|
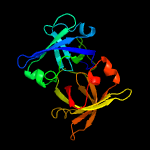 |
100.0 |
26 |
PDB header:luminescent protein
Chain: B: PDB Molecule:lumazine protein;
PDBTitle: crystal structure of lump complexed with riboflavin
|
|
|
|
| 5 | c3ddyA_
|
|
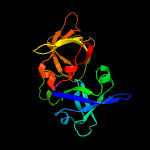 |
100.0 |
28 |
PDB header:luminescent protein
Chain: A: PDB Molecule:lumazine protein;
PDBTitle: structure of lumazine protein, an optical transponder of luminescent2 bacteria
|
|
|
|
| 6 | d1kzla2
|
|
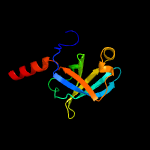 |
100.0 |
36 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
|
|
|
| 7 | d1i8da2
|
|
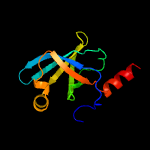 |
100.0 |
37 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
|
|
|
| 8 | c1i18B_
|
|
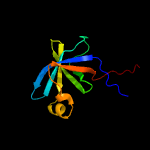 |
100.0 |
36 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: solution structure of the n-terminal domain of riboflavin synthase2 from e. coli
|
|
|
|
| 9 | c1hzeB_
|
|
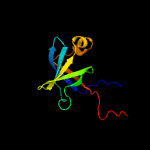 |
100.0 |
36 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: solution structure of the n-terminal domain of riboflavin synthase2 from e. coli
|
|
|
|
| 10 | d1i8da1
|
|
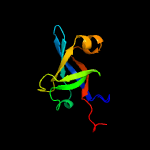 |
100.0 |
35 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
|
|
|
| 11 | c1pkvA_
|
|
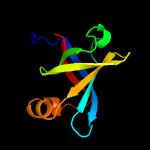 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
|
|
|
| 12 | d1kzla1
|
|
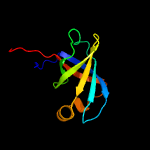 |
100.0 |
43 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
|
|
|
| 13 | c1pkvB_
|
|
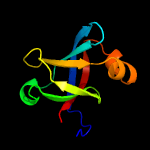 |
100.0 |
36 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
|
|
|
| 14 | c5opjA_
|
|
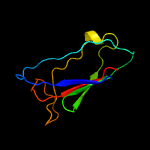 |
79.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rhamnogalacturonan lyase;
PDBTitle: rhamnogalacturonan lyase
|
|
|
|
| 15 | d1a3xa1
|
|
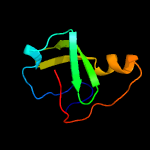 |
69.5 |
25 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
|
| 16 | d1pkla1
|
|
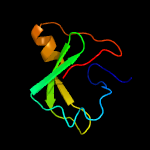 |
69.1 |
18 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
|
| 17 | c5mqoA_
|
|
 |
66.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:non-reducing end beta-l-arabinofuranosidase;
PDBTitle: glycoside hydrolase bt_1003
|
|
|
|
| 18 | c4qk0C_
|
|
 |
66.1 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:gh127 beta-l-arabinofuranoside;
PDBTitle: crystal structure of ara127n-se, a gh127 beta-l-arabinofuranosidase2 from geobacillus stearothermophilus t6
|
|
|
|
| 19 | d2g50a1
|
|
 |
57.6 |
29 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
|
| 20 | c6ex6A_
|
|
 |
53.2 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:six-hairpin glycosidase;
PDBTitle: the gh127, beta-arabinofuranosidase, bt3674
|
|
|
|
| 21 | c1vw4N_ |
|
not modelled |
48.2 |
16 |
PDB header:ribosome
Chain: N: PDB Molecule:54s ribosomal protein l49, mitochondrial;
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
|
|
|
| 22 | d1ep3b1 |
|
not modelled |
47.2 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 23 | c3o44G_ |
|
not modelled |
41.0 |
35 |
PDB header:toxin
Chain: G: PDB Molecule:hemolysin;
PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya) heptameric2 pore
|
|
|
| 24 | d1krha1 |
|
not modelled |
40.5 |
14 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 25 | d1liua1 |
|
not modelled |
37.7 |
22 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
| 26 | c1xezA_ |
|
not modelled |
35.0 |
35 |
PDB header:toxin
Chain: A: PDB Molecule:hemolysin;
PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya)2 pro-toxin with octylglucoside bound
|
|
|
| 27 | c6cqoH_ |
|
not modelled |
35.0 |
16 |
PDB header:dna binding protein
Chain: H: PDB Molecule:single-stranded dna-binding protein rim1, mitochondrial;
PDBTitle: crystal structure of mitochondrial single-stranded dna binding2 proteins from s. cerevisiae (semet labeled), rim1 (form2)
|
|
|
| 28 | c3wkxA_ |
|
not modelled |
33.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:non-reducing end beta-l-arabinofuranosidase;
PDBTitle: crystal structure of gh127 beta-l-arabinofuranosidase hypba1 from2 bifidobacterium longum arabinose complex form
|
|
|
| 29 | d1e0ta1 |
|
not modelled |
32.8 |
24 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
| 30 | d1pkma1 |
|
not modelled |
31.1 |
22 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
|
|
| 31 | c3ca9A_ |
|
not modelled |
30.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxyuridine triphosphatase;
PDBTitle: evolution of chlorella virus dutpase
|
|
|
| 32 | c4pwuC_ |
|
not modelled |
29.7 |
13 |
PDB header:signaling protein
Chain: C: PDB Molecule:modulator protein mzra;
PDBTitle: crystal structure of a modulator protein mzra (kpn_03524) from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.45 a3 resolution
|
|
|
| 33 | d1f7da_ |
|
not modelled |
29.5 |
15 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
|
|
| 34 | c5xrwA_ |
|
not modelled |
29.3 |
18 |
PDB header:motor protein
Chain: A: PDB Molecule:flin;
PDBTitle: crystal structure of flagellar motor switch complex from h. pylori
|
|
|
| 35 | c3c3iA_ |
|
not modelled |
28.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxyuridine triphosphatase;
PDBTitle: evolution of chlorella virus dutpase
|
|
|
| 36 | d1duna_ |
|
not modelled |
28.0 |
15 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
|
|
| 37 | d1f7ra_ |
|
not modelled |
27.7 |
20 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
|
|
| 38 | c5vjyC_ |
|
not modelled |
27.4 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:dutp pyrophosphatase;
PDBTitle: crystal structure of dutp pyrophosphatase protein, from naegleria2 fowleri
|
|
|
| 39 | c2d4nA_ |
|
not modelled |
27.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:du;
PDBTitle: crystal structure of m-pmv dutpase complexed with dupnpp, substrate2 analogue
|
|
|
| 40 | d1zbfa1 |
|
not modelled |
26.4 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
|
|
| 41 | d1tvca1 |
|
not modelled |
24.4 |
19 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 42 | d1fdra1 |
|
not modelled |
22.4 |
13 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 43 | c2zp2B_ |
|
not modelled |
22.2 |
18 |
PDB header:transferase inhibitor
Chain: B: PDB Molecule:kinase a inhibitor;
PDBTitle: c-terminal domain of kipi from bacillus subtilis
|
|
|
| 44 | c2okdB_ |
|
not modelled |
22.1 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:deoxyuridine 5'-triphosphate nucleotidohydrolase;
PDBTitle: high resolution crystal structures of vaccinia virus dutpase
|
|
|
| 45 | c1v0eB_ |
|
not modelled |
21.5 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:endo-alpha-sialidase;
PDBTitle: endosialidase of bacteriophage k1f
|
|
|
| 46 | c4or1B_ |
|
not modelled |
20.1 |
50 |
PDB header:cell adhesion
Chain: B: PDB Molecule:invasin homolog aafb, major fimbrial subunit of aggregative
PDBTitle: structure and mechanism of fibronectin binding and biofilm formation2 of enteroaggregative escherischia coli aaf fimbriae
|
|
|
| 47 | d1q5uz_ |
|
not modelled |
19.6 |
17 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
|
|
| 48 | c3lqwA_ |
|
not modelled |
18.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxyuridine 5'-triphosphate nucleotidohydrolase;
PDBTitle: crystal structure of deoxyuridine 5-triphosphate2 nucleotidohydrolase from entamoeba histolytica
|
|
|
| 49 | c5hbzE_ |
|
not modelled |
18.7 |
32 |
PDB header:hydrolase
Chain: E: PDB Molecule:non-structural protein 11;
PDBTitle: structure of eav nsp11 k170a mutant at 3.10a
|
|
|
| 50 | c1zeqX_ |
|
not modelled |
18.7 |
26 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
|
|
| 51 | c4bwdA_ |
|
not modelled |
18.3 |
38 |
PDB header:structural protein
Chain: A: PDB Molecule:short coiled-coil protein;
PDBTitle: human short coiled coil protein
|
|
|
| 52 | c3ehwA_ |
|
not modelled |
18.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dutp pyrophosphatase;
PDBTitle: human dutpase in complex with alpha,beta-imido-dutp and mg2+:2 visualization of the full-length c-termini in all monomers and3 suggestion for an additional metal ion binding site
|
|
|
| 53 | c2wg6L_ |
|
not modelled |
18.1 |
16 |
PDB header:transcription,hydrolase
Chain: L: PDB Molecule:general control protein gcn4, proteasome-activating
PDBTitle: proteasome-activating nucleotidase (pan) n-domain (57-134) from2 archaeoglobus fulgidus fused to gcn4, p61a mutant
|
|
|
| 54 | d1lqsl_ |
|
not modelled |
18.0 |
25 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Interferons/interleukin-10 (IL-10) |
|
|
| 55 | c5mmiS_ |
|
not modelled |
17.3 |
15 |
PDB header:ribosome
Chain: S: PDB Molecule:50s ribosomal protein l21, chloroplastic;
PDBTitle: structure of the large subunit of the chloroplast ribosome
|
|
|
| 56 | d1vlka_ |
|
not modelled |
17.2 |
31 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Interferons/interleukin-10 (IL-10) |
|
|
| 57 | c3nrfA_ |
|
not modelled |
17.1 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:apag protein;
PDBTitle: crystal structure of an apag protein (pa1934) from pseudomonas2 aeruginosa pao1 at 1.50 a resolution
|
|
|
| 58 | c3qtgA_ |
|
not modelled |
16.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from pyrobaculum aerophilum
|
|
|
| 59 | d1lk3a_ |
|
not modelled |
16.9 |
31 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Interferons/interleukin-10 (IL-10) |
|
|
| 60 | c3so2A_ |
|
not modelled |
15.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: chlorella dutpase
|
|
|
| 61 | c3h43F_ |
|
not modelled |
15.5 |
27 |
PDB header:hydrolase
Chain: F: PDB Molecule:proteasome-activating nucleotidase;
PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
|
|
|
| 62 | c3mmlD_ |
|
not modelled |
14.7 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:allophanate hydrolase subunit 1;
PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
|
|
|
| 63 | c5o57A_ |
|
not modelled |
14.7 |
40 |
PDB header:signaling protein
Chain: A: PDB Molecule:dickkopf-related protein 4;
PDBTitle: solution structure of the n-terminal region of dkk4
|
|
|
| 64 | d1vqoq1 |
|
not modelled |
14.6 |
24 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
|
|
| 65 | c3j21R_ |
|
not modelled |
14.4 |
29 |
PDB header:ribosome
Chain: R: PDB Molecule:50s ribosomal protein l21e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
|
|
|
| 66 | d1k8kg_ |
|
not modelled |
14.2 |
29 |
Fold:alpha-alpha superhelix
Superfamily:Arp2/3 complex 16 kDa subunit ARPC5
Family:Arp2/3 complex 16 kDa subunit ARPC5 |
|
|
| 67 | c2izpB_ |
|
not modelled |
13.8 |
31 |
PDB header:toxin
Chain: B: PDB Molecule:putative membrane antigen;
PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.
|
|
|
| 68 | c3mbqC_ |
|
not modelled |
13.4 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxyuridine 5'-triphosphate nucleotidohydrolase;
PDBTitle: crystal structure of deoxyuridine 5-triphosphate nucleotidohydrolase2 from brucella melitensis, orthorhombic crystal form
|
|
|
| 69 | c5mlcT_ |
|
not modelled |
13.3 |
15 |
PDB header:ribosome
Chain: T: PDB Molecule:50s ribosomal protein l21, chloroplastic;
PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
|
|
|
| 70 | c2lmcB_ |
|
not modelled |
13.1 |
6 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: structure of t7 transcription factor gp2-e. coli rnap jaw domain2 complex
|
|
|
| 71 | d2phcb1 |
|
not modelled |
12.8 |
18 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:PH0987 C-terminal domain-like |
|
|
| 72 | c3uiyA_ |
|
not modelled |
12.8 |
19 |
PDB header:structural protein
Chain: A: PDB Molecule:chimera protein of sefd and sefa;
PDBTitle: crystal structure of sefd_dsca in h2o
|
|
|
| 73 | d2izpa1 |
|
not modelled |
12.7 |
31 |
Fold:IpaD-like
Superfamily:IpaD-like
Family:IpaD-like |
|
|
| 74 | c2jz2A_ |
|
not modelled |
12.6 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ssl0352 protein;
PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
|
|
|
| 75 | c2zkrq_ |
|
not modelled |
12.6 |
18 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:rna expansion segment es31 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
|
|
|
| 76 | c4ce4V_ |
|
not modelled |
12.1 |
17 |
PDB header:ribosome
Chain: V: PDB Molecule:mrpl21;
PDBTitle: 39s large subunit of the porcine mitochondrial ribosome
|
|
|
| 77 | c4bq7D_ |
|
not modelled |
12.1 |
27 |
PDB header:cell adhesion
Chain: D: PDB Molecule:rgm domain family member b;
PDBTitle: crystal structure of the rgmb-neo1 complex form 2
|
|
|
| 78 | d3ehwa1 |
|
not modelled |
12.0 |
17 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
|
|
| 79 | c3f4fB_ |
|
not modelled |
11.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:deoxyuridine 5'-triphosphate nucleotidohydrolase;
PDBTitle: crystal structure of dut1p, a dutpase from saccharomyces cerevisiae
|
|
|
| 80 | c3mtvA_ |
|
not modelled |
11.7 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:papain-like cysteine protease;
PDBTitle: the crystal structure of the prrsv nonstructural protein nsp1
|
|
|
| 81 | d3bzka4 |
|
not modelled |
11.5 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
|
|
| 82 | d1o6aa_ |
|
not modelled |
11.4 |
18 |
Fold:Surface presentation of antigens (SPOA)
Superfamily:Surface presentation of antigens (SPOA)
Family:Surface presentation of antigens (SPOA) |
|
|
| 83 | c5vgzC_ |
|
not modelled |
11.3 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:26s proteasome regulatory subunit 8;
PDBTitle: conformational landscape of the p28-bound human proteasome regulatory2 particle
|
|
|
| 84 | c1s1iQ_ |
|
not modelled |
11.1 |
29 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l21-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
|
|
|
| 85 | d2ilka_ |
|
not modelled |
10.8 |
31 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Interferons/interleukin-10 (IL-10) |
|
|
| 86 | c3zleI_ |
|
not modelled |
10.6 |
17 |
PDB header:membrane protein
Chain: I: PDB Molecule:apical membrane antigen 1;
PDBTitle: crystal structure of toxoplasma gondii sporozoite ama1
|
|
|
| 87 | d1a8pa1 |
|
not modelled |
10.5 |
12 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 88 | c5vgzF_ |
|
not modelled |
10.3 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:26s proteasome regulatory subunit 6a;
PDBTitle: conformational landscape of the p28-bound human proteasome regulatory2 particle
|
|
|
| 89 | c5da1B_ |
|
not modelled |
10.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoribonuclease;
PDBTitle: a dimerization-dependent mechanism drives prrsv nsp11 functions as a2 beta interferon antagonist and endoribonuclease
|
|
|
| 90 | c3kf8D_ |
|
not modelled |
10.3 |
24 |
PDB header:structural protein
Chain: D: PDB Molecule:protein ten1;
PDBTitle: crystal structure of c. tropicalis stn1-ten1 complex
|
|
|
| 91 | c4yxaB_ |
|
not modelled |
10.2 |
16 |
PDB header:protein transport
Chain: B: PDB Molecule:surface presentation of antigens protein spao;
PDBTitle: complex of spao(spoa1,2 semet) and orgb(apar)::t4lysozyme fusion2 protein
|
|
|
| 92 | d1vkpa_ |
|
not modelled |
10.1 |
25 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
|
|
| 93 | c2l55A_ |
|
not modelled |
9.9 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
|
|
| 94 | c2p9oB_ |
|
not modelled |
9.9 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:dutp pyrophosphatase-like protein;
PDBTitle: structure of dutpase from arabidopsis thaliana
|
|
|
| 95 | c3e0eA_ |
|
not modelled |
9.8 |
20 |
PDB header:replication
Chain: A: PDB Molecule:replication protein a;
PDBTitle: crystal structure of a domain of replication protein a from2 methanococcus maripaludis. northeast structural genomics3 targe mrr110b
|
|
|
| 96 | c3voqA_ |
|
not modelled |
9.4 |
33 |
PDB header:membrane protein
Chain: A: PDB Molecule:target of rapamycin complex 2 subunit mapkap1;
PDBTitle: crystal structure of the pleckstrin homology domain of human sin1, a2 torc2 subunit
|
|
|
| 97 | c3iz5U_ |
|
not modelled |
9.4 |
35 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein l21 (l21e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
|
|
| 98 | c3izcU_ |
|
not modelled |
9.2 |
29 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein rpl21 (l21e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 99 | d1r9fa_ |
|
not modelled |
9.1 |
55 |
Fold:Tombusvirus P19 core protein, VP19
Superfamily:Tombusvirus P19 core protein, VP19
Family:Tombusvirus P19 core protein, VP19 |
|
|






























































































































































