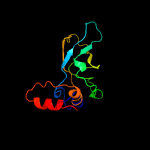
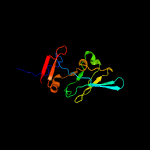
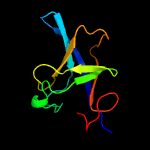
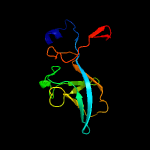
1 c3anuA_
100.0
27
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
2 c4v15B_
100.0
31
PDB header: lyaseChain: B: PDB Molecule: d-threonine aldolase;PDBTitle: crystal structure of d-threonine aldolase from alcaligenes2 xylosoxidans
3 c3wqgB_
100.0
25
PDB header: lyaseChain: B: PDB Molecule: d-threo-3-hydroxyaspartate dehydratase;PDBTitle: d-threo-3-hydroxyaspartate dehydratase c353a mutant in the metal-free2 form
4 c3llxA_
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: predicted amino acid aldolase or racemase;PDBTitle: crystal structure of an ala racemase-like protein (il1761) from2 idiomarina loihiensis at 1.50 a resolution
5 c3gwqB_
100.0
22
PDB header: lyaseChain: B: PDB Molecule: d-serine deaminase;PDBTitle: crystal structure of a putative d-serine deaminase (bxe_a4060) from2 burkholderia xenovorans lb400 at 2.00 a resolution
6 c2dy3B_
97.6
14
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from corynebacterium glutamicum
7 c5irpA_
97.5
17
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 2;PDBTitle: crystal structure of the alanine racemase bsu17640 from bacillus2 subtilis
8 c4y2wA_
97.2
20
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 1;PDBTitle: crystal structure of a thermostable alanine racemase from2 thermoanaerobacter tengcongensis mb4
9 c1xfcB_
96.9
13
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: the 1.9 a crystal structure of alanine racemase from mycobacterium2 tuberculosis contains a conserved entryway into the active site
10 c4tloB_
96.9
14
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: alanine racemase from acinetobacter baumannii
11 c3oo2B_
95.9
23
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
12 c5yycC_
95.7
13
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from bacillus pseudofirmus (of4)
13 c2yxxA_
94.8
17
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
14 c4kbxA_
94.3
9
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yhfx;PDBTitle: crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx2 from escherichia coli
15 c4eclA_
93.8
16
PDB header: isomeraseChain: A: PDB Molecule: serine racemase;PDBTitle: crystal structure of the cytoplasmic domain of vancomycin resistance2 serine racemase vantg
16 c1vftA_
92.7
13
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of l-cycloserine-bound form of alanine2 racemase from d-cycloserine-producing streptomyces3 lavendulae
17 c4lusD_
91.6
11
PDB header: isomeraseChain: D: PDB Molecule: alanine racemase;PDBTitle: alanine racemase [clostridium difficile 630]
18 c3kw3B_
89.0
21
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from bartonella henselae with2 covalently bound pyridoxal phosphate
19 c2rjgC_
88.8
14
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of biosynthetic alaine racemase from escherichia2 coli
20 c4bf5A_
88.7
17
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: structure of broad spectrum racemase from aeromonas hydrophila
21 c3mt1B_
not modelled
82.9
15
PDB header: lyaseChain: B: PDB Molecule: putative carboxynorspermidine decarboxylase protein;PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
22 c2odoC_
not modelled
81.3
17
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of pseudomonas fluorescens alanine racemase
23 c4dzaA_
not modelled
81.3
15
PDB header: isomeraseChain: A: PDB Molecule: lysine racemase;PDBTitle: crystal structure of a lysine racemase within internal aldimine2 linkage
24 c5bwaA_
not modelled
80.8
15
PDB header: lyase/lyase inhibitorChain: A: PDB Molecule: ornithine decarboxylase;PDBTitle: crystal structure of odc-plp-az1 ternary complex
25 c1niuA_
not modelled
79.8
16
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: alanine racemase with bound inhibitor derived from l-2 cycloserine
26 c3co8B_
not modelled
79.5
13
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from oenococcus oeni
27 c2qghA_
not modelled
75.9
14
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase from helicobacter2 pylori complexed with l-lysine
28 c4beqA_
not modelled
73.3
13
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 2;PDBTitle: structure of vibrio cholerae broad spectrum racemase double2 mutant r173a, n174a
29 c5gjmB_
not modelled
69.5
14
PDB header: lyaseChain: B: PDB Molecule: lysine/ornithine decarboxylase;PDBTitle: crystal strcuture of lysine decarboxylase from selenomonas ruminantium2 in c2 space group
30 c1tufA_
not modelled
68.3
11
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase from m.2 jannaschi
31 c3n2bD_
not modelled
66.4
12
PDB header: lyaseChain: D: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
32 c3oo2A_
not modelled
65.8
24
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
33 c2p3eA_
not modelled
64.4
12
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of aq1208 from aquifex aeolicus
34 c4fs9B_
not modelled
63.9
19
PDB header: isomeraseChain: B: PDB Molecule: broad specificity amino acid racemase;PDBTitle: complex structure of a broad specificity amino acid racemase (bar)2 within the reactive intermediate
35 c5zl6A_
not modelled
58.2
17
PDB header: isomeraseChain: A: PDB Molecule: histidine racemase;PDBTitle: histidine racemase from leuconostoc mesenteroides subsp. sake nbrc2 102480
36 c6a2fB_
not modelled
57.2
21
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase, biosynthetic;PDBTitle: crystal structure of biosynthetic alanine racemase from pseudomonas2 aeruginosa
37 c5x7nA_
not modelled
55.2
20
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal strcuture of meso-diaminopimelate decarboxylase (dapdc) from2 corynebacterium glutamicum
38 c3cosD_
not modelled
48.3
7
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase 4;PDBTitle: crystal structure of human class ii alcohol dehydrogenase (adh4) in2 complex with nad and zn
39 c1ma0B_
not modelled
44.7
7
PDB header: oxidoreductaseChain: B: PDB Molecule: glutathione-dependent formaldehyde dehydrogenase;PDBTitle: ternary complex of human glutathione-dependent formaldehyde2 dehydrogenase with nad+ and dodecanoic acid
40 c2pljA_
not modelled
41.3
15
PDB header: lyaseChain: A: PDB Molecule: lysine/ornithine decarboxylase;PDBTitle: crystal structure of lysine/ornithine decarboxylase complexed with2 putrescine from vibrio vulnificus
41 c2ouiB_
not modelled
39.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nadp-dependent alcohol dehydrogenase;PDBTitle: d275p mutant of alcohol dehydrogenase from protozoa entamoeba2 histolytica
42 c3ukoA_
not modelled
38.1
7
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase class-3;PDBTitle: crystal structure of s-nitrosoglutathione reductase from arabidopsis2 thaliana, complex with nadh
43 c3mubB_
not modelled
36.2
26
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: the crystal structure of alanine racemase from streptococcus2 pneumoniae
44 d2toda1
not modelled
35.7
18
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like
45 c2nvaH_
not modelled
35.5
14
PDB header: lyaseChain: H: PDB Molecule: arginine decarboxylase, a207r protein;PDBTitle: the x-ray crystal structure of the paramecium bursaria chlorella virus2 arginine decarboxylase bound to agmatine
46 c4rquA_
not modelled
35.0
11
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: alcohol dehydrogenase crystal structure in complex with nad
47 c4gi2B_
not modelled
34.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: crotonyl-coa carboxylase/reductase;PDBTitle: crotonyl-coa carboxylase/reductase
48 c1knwA_
not modelled
32.9
18
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase
49 c5ylnB_
not modelled
32.8
8
PDB header: oxidoreductaseChain: B: PDB Molecule: alcohol dehydrogenase, zinc-containing;PDBTitle: zinc dependent alcohol dehydrogenase 2 from streptococcus pneumonia -2 apo form
50 c1r37B_
not modelled
31.2
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nad-dependent alcohol dehydrogenase;PDBTitle: alcohol dehydrogenase from sulfolobus solfataricus2 complexed with nad(h) and 2-ethoxyethanol
51 c1e3jA_
not modelled
30.8
24
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp(h)-dependent ketose reductase;PDBTitle: ketose reductase (sorbitol dehydrogenase) from silverleaf2 whitefly
52 c5vm2A_
not modelled
30.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: crystal structure of eck1772, an oxidoreductase/dehydrogenase of2 unknown specificity involved in membrane biogenesis from escherichia3 coli
53 c4cpdA_
not modelled
29.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: alcohol dehydrogenase tadh from thermus sp. atn1
54 c5kiaA_
not modelled
29.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: l-threonine 3-dehydrogenase;PDBTitle: crystal structure of l-threonine 3-dehydrogenase from burkholderia2 thailandensis
55 c1kolA_
not modelled
28.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: formaldehyde dehydrogenase;PDBTitle: crystal structure of formaldehyde dehydrogenase
56 c5k1sD_
not modelled
26.1
18
PDB header: oxidoreductaseChain: D: PDB Molecule: oxidoreductase, zinc-binding dehydrogenase family;PDBTitle: crystal structure of aibc
57 c2j66A_
not modelled
25.6
13
PDB header: lyaseChain: A: PDB Molecule: btrk;PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
58 c4ilkB_
not modelled
24.9
21
PDB header: oxidoreductaseChain: B: PDB Molecule: starvation sensing protein rspb;PDBTitle: crystal structure of short chain alcohol dehydrogenase (rspb) from e.2 coli cft073 (efi target efi-506413) complexed with cofactor nadh
59 c4gkvC_
not modelled
24.8
12
PDB header: oxidoreductaseChain: C: PDB Molecule: alcohol dehydrogenase, propanol-preferring;PDBTitle: structure of escherichia coli adhp (ethanol-inducible dehydrogenase)2 with bound nad
60 c2eihA_
not modelled
24.6
12
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: crystal structure of nad-dependent alcohol dehydrogenase
61 c3hurA_
not modelled
22.8
17
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from oenococcus oeni
62 c2dphA_
not modelled
22.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: formaldehyde dismutase;PDBTitle: crystal structure of formaldehyde dismutase
63 c4a2cB_
not modelled
21.9
12
PDB header: oxidoreductaseChain: B: PDB Molecule: galactitol-1-phosphate 5-dehydrogenase;PDBTitle: crystal structure of galactitol-1-phosphate dehydrogenase from2 escherichia coli
64 c4eezB_
not modelled
21.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: alcohol dehydrogenase 1;PDBTitle: crystal structure of lactococcus lactis alcohol dehydrogenase variant2 re1
65 c3krtC_
not modelled
20.7
8
PDB header: oxidoreductaseChain: C: PDB Molecule: crotonyl coa reductase;PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
66 c1h2bA_
not modelled
20.7
8
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: crystal structure of the alcohol dehydrogenase from the2 hyperthermophilic archaeon aeropyrum pernix at 1.65a resolution
67 c3m6iA_
not modelled
20.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arabinitol 4-dehydrogenase;PDBTitle: l-arabinitol 4-dehydrogenase
68 c2dfvB_
not modelled
19.8
21
PDB header: oxidoreductaseChain: B: PDB Molecule: probable l-threonine 3-dehydrogenase;PDBTitle: hyperthermophilic threonine dehydrogenase from pyrococcus horikoshii
69 c2ejvA_
not modelled
19.3
20
PDB header: oxidoreductaseChain: A: PDB Molecule: l-threonine 3-dehydrogenase;PDBTitle: crystal structure of threonine 3-dehydrogenase complexed with nad+
70 c1cdoB_
not modelled
18.1
7
PDB header: oxidoreductase (ch-oh(d)-nad(a))Chain: B: PDB Molecule: alcohol dehydrogenase;PDBTitle: alcohol dehydrogenase (e.c.1.1.1.1) (ee isozyme) complexed with2 nicotinamide adenine dinucleotide (nad), and zinc
71 c3qf4B_
not modelled
17.9
23
PDB header: transport proteinChain: B: PDB Molecule: uncharacterized abc transporter atp-binding proteinPDBTitle: crystal structure of a heterodimeric abc transporter in its inward-2 facing conformation
72 c1hf3A_
not modelled
17.8
8
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase e chain;PDBTitle: atomic x-ray structure of liver alcohol dehydrogenase containing2 cadmium and a hydroxide adduct to nadh
73 c2dxcG_
not modelled
17.4
20
PDB header: hydrolaseChain: G: PDB Molecule: thiocyanate hydrolase subunit alpha;PDBTitle: recombinant thiocyanate hydrolase, fully-matured form
74 c3b5xB_
not modelled
17.4
23
PDB header: membrane proteinChain: B: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: crystal structure of msba from vibrio cholerae
75 d2nqra1
not modelled
17.1
22
Fold: beta-clipSuperfamily: MoeA C-terminal domain-likeFamily: MoeA C-terminal domain-like
76 c2qf4A_
not modelled
16.6
35
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
77 c3qz9D_
not modelled
15.8
17
PDB header: lyaseChain: D: PDB Molecule: co-type nitrile hydratase beta subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
78 c2j5uB_
not modelled
15.7
27
PDB header: cell shape regulationChain: B: PDB Molecule: mrec protein;PDBTitle: mrec lysteria monocytogenes
79 c1kevB_
not modelled
15.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nadp-dependent alcohol dehydrogenase;PDBTitle: structure of nadp-dependent alcohol dehydrogenase
80 c4ejmA_
not modelled
15.3
24
PDB header: oxidoreductaseChain: A: PDB Molecule: putative zinc-binding dehydrogenase;PDBTitle: crystal structure of a putative zinc-binding dehydrogenase (target2 psi-012003) from sinorhizobium meliloti 1021 bound to nadp
81 c1pl6A_
not modelled
14.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: sorbitol dehydrogenase;PDBTitle: human sdh/nadh/inhibitor complex
82 c5mkkA_
not modelled
14.7
28
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance abc transporter atp-binding andPDBTitle: crystal structure of the heterodimeric abc transporter tmrab, a2 homolog of the antigen translocation complex tap
83 c1lluD_
not modelled
14.3
8
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase;PDBTitle: the ternary complex of pseudomonas aeruginosa alcohol2 dehydrogenase with its coenzyme and weak substrate
84 c2cf5A_
not modelled
14.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: cinnamyl alcohol dehydrogenase;PDBTitle: crystal structures of the arabidopsis cinnamyl alcohol2 dehydrogenases, atcad5
85 c1s1iQ_
not modelled
13.5
20
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l21-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
86 d2ix0a3
not modelled
13.4
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
87 c1p0fA_
not modelled
12.9
7
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp-dependent alcohol dehydrogenase;PDBTitle: crystal structure of the binary complex: nadp(h)-dependent vertebrate2 alcohol dehydrogenase (adh8) with the cofactor nadp
88 c3j21R_
not modelled
12.7
12
PDB header: ribosomeChain: R: PDB Molecule: 50s ribosomal protein l21e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
89 d1vqoq1
not modelled
12.5
8
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e
90 c5ochH_
not modelled
12.2
16
PDB header: hydrolaseChain: H: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
91 c2yl4A_
not modelled
12.0
19
PDB header: membrane proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter, abcb10
92 c2lo4A_
not modelled
11.9
36
PDB header: protein transportChain: A: PDB Molecule: optineurin;PDBTitle: nmr solution structure of optineurin zinc-finger domain
93 d1ugpb_
not modelled
11.8
17
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain
94 d1uufa1
not modelled
11.7
16
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain
95 d1kola1
not modelled
11.6
20
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain
96 c4jbiB_
not modelled
11.3
12
PDB header: oxidoreductaseChain: B: PDB Molecule: alcohol dehydrogenase (zinc);PDBTitle: 2.35a resolution structure of nadph bound thermostable alcohol2 dehydrogenase from pyrobaculum aerophilum
97 c2vwpA_
not modelled
11.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: glucose dehydrogenase;PDBTitle: haloferax mediterranei glucose dehydrogenase in complex with nadph and2 zn.
98 c3qf4A_
not modelled
10.8
16
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, atp-binding protein;PDBTitle: crystal structure of a heterodimeric abc transporter in its inward-2 facing conformation
99 c3iz5U_
not modelled
10.7
14
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome