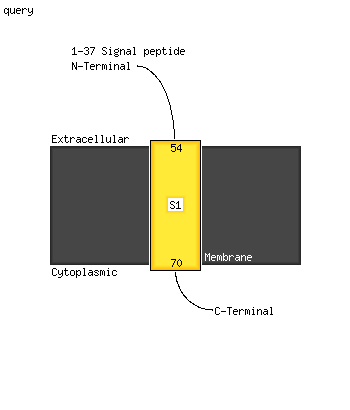
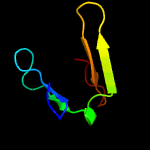
1 d1ef1a2
47.9
20
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
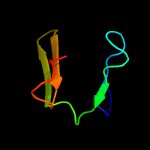
2 d1gg3a2
39.6
26
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
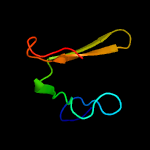
3 d1e5wa2
38.4
18
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
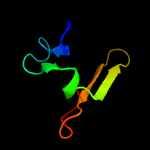
4 d1j19a2
36.7
18
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
5 d1isna2
34.2
14
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
6 c1e5wA_
31.7
18
PDB header: membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: structure of isolated ferm domain and first long helix of moesin
7 d3bzka5
26.8
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like
8 c6adqP_
23.7
18
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
9 d1iwga2
20.9
20
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
10 c2kl4A_
18.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2032 protein;PDBTitle: nmr structure of the protein nb7804a
11 c2i1jA_
17.1
26
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda at 2.1 angstroms resolution
12 d1h4ra2
15.9
17
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
13 c4gioA_
15.4
9
PDB header: unknown functionChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of campylobacter jejuni cj0090
14 c6ijjK_
12.2
36
PDB header: membrane proteinChain: K: PDB Molecule: psak;PDBTitle: photosystem i of chlamydomonas reinhardtii
15 c6d2qA_
11.9
27
PDB header: signaling proteinChain: A: PDB Molecule: ferm, rhogef (arhgef) and pleckstrin domain protein 1PDBTitle: crystal structure of the ferm domain of zebrafish farp1
16 c6igzK_
11.7
24
PDB header: plant proteinChain: K: PDB Molecule: psak;PDBTitle: structure of psi-lhci
17 d2zpya2
11.6
20
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Third domain of FERM
18 c2i1kA_
10.8
27
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda reveals the coiled-coil domain at2 3.0 angstrom resolution
19 c6odih_
10.3
24
PDB header: translocaseChain: H: PDB Molecule: PDBTitle: structure of cagy from a cryo-em reconstruction of a t4ss
20 c5n76C_
9.8
22
PDB header: nickel-binding proteinChain: C: PDB Molecule: coot;PDBTitle: crystal structure of the apo-form of the co dehydrogenase accessory2 protein coot from rhodospirillum rubrum
21 c3u8zD_
not modelled
8.4
17
PDB header: signaling proteinChain: D: PDB Molecule: merlin;PDBTitle: human merlin ferm domain
22 c6ijoG_
not modelled
8.1
36
PDB header: photosynthesisChain: G: PDB Molecule: psag;PDBTitle: photosystem i of chlamydomonas reinhardtii
23 d2gf3a2
not modelled
7.8
20
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: D-aminoacid oxidase-like
24 c3nqnB_
not modelled
7.7
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function. (dr_2006) from2 deinococcus radiodurans at 1.88 a resolution
25 c2emtB_
not modelled
7.7
20
PDB header: cell adhesionChain: B: PDB Molecule: radixin;PDBTitle: crystal structure analysis of the radixin ferm domain complexed with2 adhesion molecule psgl-1
26 c3lw5K_
not modelled
6.0
30
PDB header: photosynthesisChain: K: PDB Molecule: photosystem i reaction center subunit x psak;PDBTitle: improved model of plant photosystem i
27 c3uc0B_
not modelled
6.0
38
PDB header: viral protein/immune systemChain: B: PDB Molecule: envelope protein;PDBTitle: crystal structure of domain i of the envelope glycoprotein ectodomain2 from dengue virus serotype 4 in complex with the fab fragment of the3 chimpanzee monoclonal antibody 5h2
28 c2mckA_
not modelled
5.7
39
PDB header: hydrolaseChain: A: PDB Molecule: polyprotein;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
29 c5xu1A_
not modelled
5.5
16
PDB header: transport proteinChain: A: PDB Molecule: abc transporter atp-binding protein;PDBTitle: structure of a non-canonical abc transporter from streptococcus2 pneumoniae r6
30 d1ei5a2
not modelled
5.4
36
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains