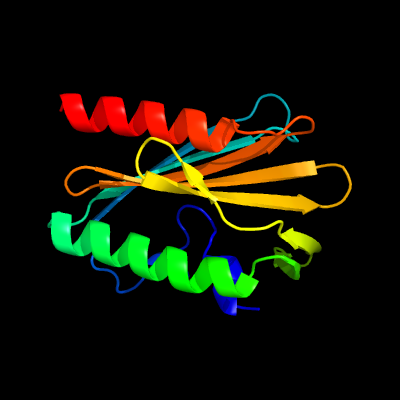
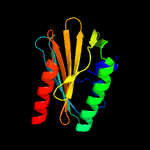
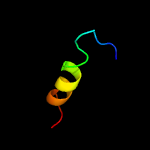
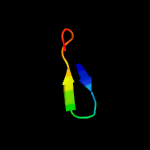
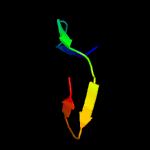
1 c4esqA_
72.6
15
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase;PDBTitle: crystal structure of the extracellular domain of pknh from2 mycobacterium tuberculosis
2 c4b9cA_
14.0
23
PDB header: carbohydrate-binding proteinChain: A: PDB Molecule: type 3a cellulose-binding domain protein;PDBTitle: biomass sensoring modules from putative rsgi-like proteins2 of clostridium thermocellum resemble family 3 carbohydrate-3 binding module of cellulosome
3 d1d1da1
13.8
32
Fold: Acyl carrier protein-likeSuperfamily: Retrovirus capsid dimerization domain-likeFamily: Retrovirus capsid protein C-terminal domain
4 c5n60Q_
12.5
46
PDB header: transferaseChain: Q: PDB Molecule: rna polymerase i-specific transcription initiation factorPDBTitle: cryo-em structure of rna polymerase i in complex with rrn3 and core2 factor (orientation i)
5 c5dudB_
12.3
60
PDB header: unknown functionChain: B: PDB Molecule: ybgj;PDBTitle: crystal structure of e. coli ybgjk
6 c6dexB_
11.4
25
PDB header: recombinationChain: B: PDB Molecule: suppressor of hydroxyurea sensitivity protein 2;PDBTitle: structure of eremothecium gossypii shu1:shu2 complex
7 c4e6nB_
11.4
18
PDB header: protein bindingChain: B: PDB Molecule: methyltransferase type 12;PDBTitle: crystal structure of bacterial pnkp-c/hen1-n heterodimer
8 c2zp2B_
10.3
55
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
9 c3pntA_
10.2
55
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: nad+-glycohydrolase;PDBTitle: crystal structure of the streptococcus pyogenes nad+ glycohydrolase2 spn in complex with ifs, the immunity factor for spn
10 c3sumA_
9.9
33
PDB header: unknown functionChain: A: PDB Molecule: cerato-platanin-like protein;PDBTitle: crystal structure of cerato-platanin 5 from m. perniciosa (mpcp5)
11 d1iyca_
9.8
100
Fold: Invertebrate chitin-binding proteinsSuperfamily: Invertebrate chitin-binding proteinsFamily: Antifungal peptide scarabaecin
12 c3mmlD_
9.5
60
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
13 c6nd1D_
8.7
47
PDB header: protein transportChain: D: PDB Molecule: protein transport protein sbh1;PDBTitle: cryoem structure of the sec complex from yeast
14 c1vl6C_
8.4
26
PDB header: oxidoreductaseChain: C: PDB Molecule: malate oxidoreductase;PDBTitle: crystal structure of nad-dependent malic enzyme (tm0542) from2 thermotoga maritima at 2.61 a resolution
15 d1vl6a1
8.3
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
16 c3dgsA_
8.2
60
PDB header: viral proteinChain: A: PDB Molecule: coat protein a;PDBTitle: changing the determinants of protein stability from covalent to non-2 covalent interactions by in-vitro evolution: a structural and3 energetic analysis
17 c4b9pA_
8.2
43
PDB header: carbohydrate-binding proteinChain: A: PDB Molecule: type 3a cellulose-binding domain protein;PDBTitle: biomass sensoring module from putative rsgi2 protein of2 clostridium thermocellum resemble family 3 carbohydrate-3 binding module of cellulosome
18 c2ylkD_
7.7
33
PDB header: hydrolaseChain: D: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: carbohydrate-binding module cbm3b from the cellulosomal2 cellobiohydrolase 9a from clostridium thermocellum
19 c2kk4A_
7.7
41
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein af_2094;PDBTitle: solution nmr structure of protein af2094 from archaeoglobus2 fulgidus. northeast structural genomics consotium (nesg)3 target gt2
20 d2vrda1
7.6
57
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger
21 c2wwbC_
not modelled
7.2
53
PDB header: ribosomeChain: C: PDB Molecule: protein transport protein sec61 subunit beta;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the actively2 translating wheat germ 80s ribosome
22 c2rmhA_
not modelled
7.1
43
PDB header: hormoneChain: A: PDB Molecule: urocortin-3;PDBTitle: human urocortin 3
23 c2phcB_
not modelled
6.9
50
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
24 c2cosA_
not modelled
6.8
32
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase lats2;PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
25 c2outA_
not modelled
6.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mu-like prophage flumu protein gp35, proteinPDBTitle: solution structure of hi1506, a novel two domain protein2 from haemophilus influenzae
26 d1c1ka_
not modelled
6.4
25
Fold: gene 59 helicase assembly proteinSuperfamily: gene 59 helicase assembly proteinFamily: gene 59 helicase assembly protein
27 c3s3zA_
not modelled
6.0
32
PDB header: antiviral proteinChain: A: PDB Molecule: tandem cyanovirin-n dimer cvn2l10;PDBTitle: crystal structure an tandem cyanovirin-n dimer, cvn2l10
28 c4lqeA_
not modelled
6.0
39
PDB header: dna binding proteinChain: A: PDB Molecule: mepb;PDBTitle: crystal structure of mepb
29 d2phcb1
not modelled
5.9
63
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like
30 c5xynD_
not modelled
5.4
33
PDB header: dna binding proteinChain: D: PDB Molecule: suppressor of hydroxyurea sensitivity protein 2;PDBTitle: the crystal structure of csm2-psy3-shu1-shu2 complex from budding2 yeast