| 1 |
|
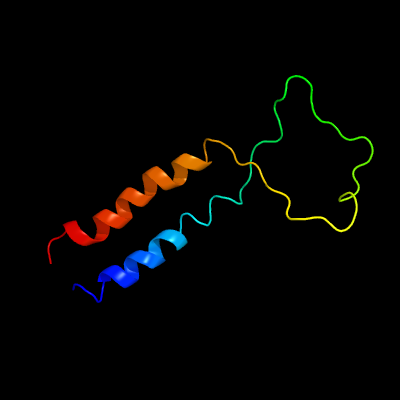
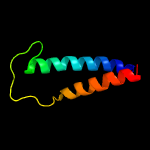
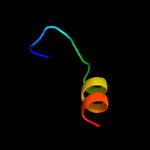
PDB 2aki chain A
Region: 1 - 77
Aligned: 77
Modelled: 77
Confidence: 98.2%
Identity: 21%
PDB header:protein transport
Chain: A: PDB Molecule:protein-export membrane protein secg;
PDBTitle: normal mode-based flexible fitted coordinates of a translocating2 secyeg protein-conducting channel into the cryo-em map of a secyeg-3 nascent chain-70s ribosome complex from e. coli
Phyre2
| 2 |
|
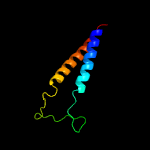
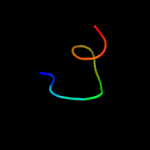
PDB 3din chain E
Region: 10 - 73
Aligned: 64
Modelled: 64
Confidence: 97.9%
Identity: 22%
PDB header:membrane protein, protein transport
Chain: E: PDB Molecule:preprotein translocase subunit secg;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 3 |
|
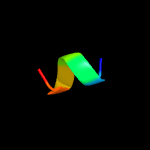
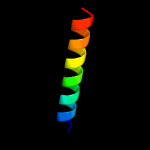
PDB 3dl8 chain F
Region: 10 - 71
Aligned: 61
Modelled: 62
Confidence: 97.5%
Identity: 28%
PDB header:protein transport
Chain: F: PDB Molecule:protein-export membrane protein secg;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and bacillus2 subtilis seca
Phyre2
| 4 |
|
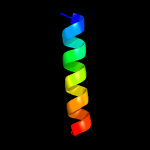
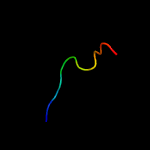
PDB 5aww chain G
Region: 3 - 75
Aligned: 71
Modelled: 73
Confidence: 87.8%
Identity: 20%
PDB header:protein transport/immune system
Chain: G: PDB Molecule:putative preprotein translocase, secg subunit;
PDBTitle: precise resting state of thermus thermophilus secyeg
Phyre2
| 5 |
|
PDB 5uz5 chain G
Region: 50 - 56
Aligned: 7
Modelled: 7
Confidence: 11.7%
Identity: 71%
PDB header:nuclear protein/rna
Chain: G: PDB Molecule:56 kda u1 small nuclear ribonucleoprotein component;
PDBTitle: s. cerevisiae u1 snrnp
Phyre2
| 6 |
|
PDB 2mxb chain A
Region: 3 - 22
Aligned: 20
Modelled: 20
Confidence: 8.5%
Identity: 40%
PDB header:membrane protein
Chain: A: PDB Molecule:erythropoietin receptor;
PDBTitle: structure of the transmembrane domain of the mouse erythropoietin2 receptor
Phyre2
| 7 |
|
PDB 1v54 chain L
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 8.1%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Family: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Phyre2
| 8 |
|
PDB 3ipr chain C
Region: 30 - 41
Aligned: 12
Modelled: 12
Confidence: 7.4%
Identity: 8%
PDB header:transferase
Chain: C: PDB Molecule:pts system, iia component;
PDBTitle: crystal structure of the enterococcus faecalis gluconate2 specific eiia phosphotransferase system component
Phyre2
| 9 |
|
PDB 1pdo chain A
Region: 30 - 40
Aligned: 11
Modelled: 11
Confidence: 7.1%
Identity: 27%
Fold: PTS system fructose IIA component-like
Superfamily: PTS system fructose IIA component-like
Family: EIIA-man component-like
Phyre2
| 10 |
|
PDB 4tkz chain A
Region: 30 - 40
Aligned: 11
Modelled: 11
Confidence: 6.9%
Identity: 36%
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein gbs1890;
PDBTitle: crystal structure of phosphotransferase system component eiia from2 streptococcus agalactiae
Phyre2
| 11 |
|
PDB 3hl6 chain B
Region: 22 - 38
Aligned: 17
Modelled: 17
Confidence: 6.9%
Identity: 24%
PDB header:unknown function
Chain: B: PDB Molecule:pathogenicity island protein;
PDBTitle: staphylococcus aureus pathogenicity island 3 orf9 protein
Phyre2
| 12 |
|
PDB 2zvf chain G
Region: 25 - 56
Aligned: 32
Modelled: 32
Confidence: 6.5%
Identity: 25%
PDB header:ligase
Chain: G: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of archaeoglobus fulgidus alanyl-trna2 synthetase c-terminal dimerization domain
Phyre2
| 13 |
|
PDB 6fmg chain C
Region: 30 - 41
Aligned: 12
Modelled: 12
Confidence: 6.4%
Identity: 25%
PDB header:transferase
Chain: C: PDB Molecule:pts system mannose-specific transporter subunit iiab;
PDBTitle: structure of the mannose transporter iia domain from streptococcus2 pneumoniae
Phyre2
| 14 |
|
PDB 2lp1 chain A
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 6.3%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:c99;
PDBTitle: the solution nmr structure of the transmembrane c-terminal domain of2 the amyloid precursor protein (c99)
Phyre2
| 15 |
|
PDB 1spf chain A
Region: 51 - 74
Aligned: 24
Modelled: 24
Confidence: 6.1%
Identity: 38%
PDB header:lipoprotein(surface film)
Chain: A: PDB Molecule:pulmonary surfactant-associated polypeptide c;
PDBTitle: the nmr structure of the pulmonary surfactant-associated2 polypeptide sp-c in an apolar solvent contains a valyl-3 rich alpha-helix
Phyre2
| 16 |
|
PDB 3lpz chain A
Region: 30 - 41
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 42%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 17 |
|
PDB 3lfh chain F
Region: 30 - 41
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 33%
PDB header:transferase
Chain: F: PDB Molecule:phosphotransferase system, mannose/fructose-specific
PDBTitle: crystal structure of manxa from thermoanaerobacter tengcongensis
Phyre2
| 18 |
|
PDB 5t3u chain A
Region: 30 - 40
Aligned: 11
Modelled: 11
Confidence: 5.6%
Identity: 36%
PDB header:transport protein
Chain: A: PDB Molecule:pts system, iia component;
PDBTitle: crystal structure of the pts iia protein associated with the fucose2 utilization operon from streptococcus pneumoniae
Phyre2